Autosomal dominant nanophthalmos and high hyperopia associated with a C-terminal frameshift variant in MYRF
- PMID: 31700225
- PMCID: PMC6817736
Autosomal dominant nanophthalmos and high hyperopia associated with a C-terminal frameshift variant in MYRF
Abstract
Purpose: Nanophthalmos is a rare subtype of microphthalmia associated with high hyperopia and an increased risk of angle-closure glaucoma. We investigated the genetic cause of nanophthalmos and high hyperopia in an autosomal dominant kindred.
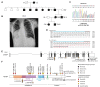
Methods: A proband with short axial length, high hyperopia, and dextrocardia was subjected to exome sequencing. Human and rodent gene expression data sets were used to investigate the expression of relevant genes.
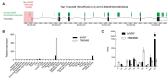
Results: We identified a segregating heterozygous frameshift variant at the 3' end of the penultimate exon of MYRF. Using Myc-MYRF chromatin immunoprecipitation data from rat oligodendrocytes, MYRF was found to bind immediately upstream of the transcriptional start site of Tmem98, a gene that itself has been implicated in autosomal dominant nanophthalmos. MYRF and TMEM98 were found to be expressed in the human retina, with a similar pattern of expression across several dissected human eye tissues.
Conclusions: C-terminal variants in MYRF, which are expected to escape nonsense-mediated decay, represent a rare cause of autosomal dominant nanophthalmos with or without dextrocardia or congenital diaphragmatic hernia.
Copyright © 2019 Molecular Vision.
Figures
References
-
- Bourne RRA, Stevens GA, White RA, Smith JL, Flaxman SR, Price H, Jonas JB, Keeffe J, Leasher J, Naidoo K, Pesudovs K, Resnikoff S, Taylor HR, Vision Loss Expert Group Causes of vision loss worldwide, 1990–2010: a systematic analysis. Lancet Glob Health. 2013;1:e339–49. - PubMed
-
- Khan AO. Recognizing posterior microphthalmos. Ophthalmology. 2006;113:718. - PubMed
-
- Khan AO. Posterior microphthalmos versus nanophthalmos. Ophthalmic Genet. 2008;29:189. - PubMed
-
- Sundin OH, Leppert GS, Silva ED, Yang J-M, Dharmaraj S, Maumenee IH, Santos LC, Parsa CF, Traboulsi EI, Broman KW, Dibernardo C, Sunness JS, Toy J, Weinberg EM. Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc Natl Acad Sci USA. 2005;102:9553–8. - PMC - PubMed
-
- Gal A, Rau I, El Matri L, Kreienkamp H-J, Fehr S, Baklouti K, Chouchane I, Li Y, Rehbein M, Fuchs J, Fledelius HC, Vilhelmsen K, Schorderet DF, Munier FL, Ostergaard E, Thompson DA, Rosenberg T. Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease. Am J Hum Genet. 2011;88:382–90. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

