FRET-based cyclic GMP biosensors measure low cGMP concentrations in cardiomyocytes and neurons
- PMID: 31701023
- PMCID: PMC6820734
- DOI: 10.1038/s42003-019-0641-x
FRET-based cyclic GMP biosensors measure low cGMP concentrations in cardiomyocytes and neurons
Abstract
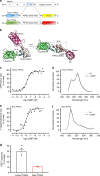
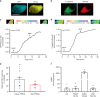
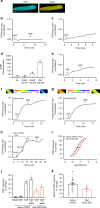
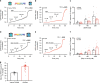
Several FRET (fluorescence resonance energy transfer)-based biosensors for intracellular detection of cyclic nucleotides have been designed in the past decade. However, few such biosensors are available for cGMP, and even fewer that detect low nanomolar cGMP concentrations. Our aim was to develop a FRET-based cGMP biosensor with high affinity for cGMP as a tool for intracellular signaling studies. We used the carboxyl-terminal cyclic nucleotide binding domain of Plasmodium falciparum cGMP-dependent protein kinase (PKG) flanked by different FRET pairs to generate two cGMP biosensors (Yellow PfPKG and Red PfPKG). Here, we report that these cGMP biosensors display high affinity for cGMP (EC50 of 23 ± 3 nM) and detect cGMP produced through soluble guanylyl cyclase and guanylyl cyclase A in stellate ganglion neurons and guanylyl cyclase B in cardiomyocytes. These biosensors are therefore optimal tools for real-time measurements of low concentrations of cGMP in living cells.
Keywords: Cell signalling; Cellular imaging; Fluorescent proteins; Membrane proteins.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors have no competing interests to declare.
Figures
Similar articles
-
Feedback control through cGMP-dependent protein kinase contributes to differential regulation and compartmentation of cGMP in rat cardiac myocytes.Circ Res. 2010 Nov 12;107(10):1232-40. doi: 10.1161/CIRCRESAHA.110.226712. Epub 2010 Sep 16. Circ Res. 2010. PMID: 20847310
-
Plasmodium falciparum Guanylyl Cyclase-Alpha and the Activity of Its Appended P4-ATPase Domain Are Essential for cGMP Synthesis and Blood-Stage Egress.mBio. 2021 Jan 26;12(1):e02694-20. doi: 10.1128/mBio.02694-20. mBio. 2021. PMID: 33500341 Free PMC article.
-
Transgenic mice for real-time visualization of cGMP in intact adult cardiomyocytes.Circ Res. 2014 Apr 11;114(8):1235-45. doi: 10.1161/CIRCRESAHA.114.302437. Epub 2014 Mar 5. Circ Res. 2014. PMID: 24599804
-
Cyclic nucleotide selectivity of protein kinase G isozymes.Protein Sci. 2021 Feb;30(2):316-327. doi: 10.1002/pro.4008. Epub 2020 Dec 10. Protein Sci. 2021. PMID: 33271627 Free PMC article. Review.
-
Cardiac cGMP Signaling in Health and Disease: Location, Location, Location.J Cardiovasc Pharmacol. 2020 May;75(5):399-409. doi: 10.1097/FJC.0000000000000802. J Cardiovasc Pharmacol. 2020. PMID: 32040034 Review.
Cited by
-
The β-cell primary cilium is an autonomous Ca2+ compartment for paracrine GABA signaling.J Cell Biol. 2023 Jan 2;222(1):e202108101. doi: 10.1083/jcb.202108101. Epub 2022 Nov 9. J Cell Biol. 2023. PMID: 36350286 Free PMC article.
-
Phosphodiesterase in heart and vessels: from physiology to diseases.Physiol Rev. 2024 Apr 1;104(2):765-834. doi: 10.1152/physrev.00015.2023. Epub 2023 Nov 16. Physiol Rev. 2024. PMID: 37971403 Free PMC article. Review.
-
CNP regulates cardiac contractility and increases cGMP near both SERCA and TnI: difference from BNP visualized by targeted cGMP biosensors.Cardiovasc Res. 2022 May 6;118(6):1506-1519. doi: 10.1093/cvr/cvab167. Cardiovasc Res. 2022. PMID: 33970224 Free PMC article.
-
CUTie2: The Attack of the Cyclic Nucleotide Sensor Clones.Front Mol Biosci. 2021 Mar 11;8:629773. doi: 10.3389/fmolb.2021.629773. eCollection 2021. Front Mol Biosci. 2021. PMID: 33778003 Free PMC article.
-
"Radical" differences between two FLIM microscopes affect interpretation of cell signaling dynamics.iScience. 2024 Jun 13;27(7):110268. doi: 10.1016/j.isci.2024.110268. eCollection 2024 Jul 19. iScience. 2024. PMID: 39036041 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

