Novel markers of human ovarian granulosa cell differentiation toward osteoblast lineage: A microarray approach
- PMID: 31702034
- PMCID: PMC6797957
- DOI: 10.3892/mmr.2019.10709
Novel markers of human ovarian granulosa cell differentiation toward osteoblast lineage: A microarray approach
Erratum in
-
[Corrigendum] Novel markers of human ovarian granulosa cell differentiation toward osteoblast lineage: A microarray approach.Mol Med Rep. 2021 Sep;24(3):669. doi: 10.3892/mmr.2021.12308. Epub 2021 Jul 23. Mol Med Rep. 2021. PMID: 34296308 Free PMC article.
Abstract
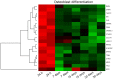
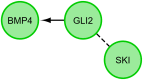
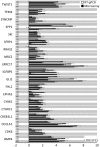
Under physiological conditions, human ovarian granulosa cells (GCs), are responsible for a number of processes associated with folliculogenesis and oogenesis. The primary functions of GCs in the individual phases of follicle growth are: Hormone production in response to follicle stimulating hormone (FSH), induction of ovarian follicle atresia through specific molecular markers and production of nexus cellular connections for communication with the oocyte. In recent years, interest in obtaining stem cells from particular tissues, including the ovary, has increased. Special attention has been paid to the novel properties of GCs during long‑term in vitro culture. It has been demonstrated that the usually recycled material in the form of follicular fluid can be a source of cells with stem‑like properties. The study group consisted of patients enrolled in the in vitro fertilization procedure. Total RNA was isolated from GCs at 4 time points (after 1, 7, 15 and 30 days of culture) and was used for microarray expression analysis (Affymetrix® Human HgU 219 Array). The expression of 22,480 transcripts was examined. The selection of significantly altered genes was based on a P‑value <0.05 and expression higher than two‑fold. The leucine rich repeat containing 17, collagen type I α1 chain, bone morphogenetic protein 4, twist family bHLH transcription factor 1, insulin like growth factor binding protein 5, GLI family zinc finger 2 and collagen triple helix repeat containing genes exhibited the highest changes in expression. Reverse‑transcription‑quantitative PCR was performed to validate the results obtained in the analysis of expression microarrays. The direction of expression changes was validated in the majority of cases. The presented results indicated that GCs have the potential of cells that can differentiate towards osteoblasts in long‑term in vitro culture conditions. Increased expression of genes associated with the osteogenesis process suggests a potential for uninduced change of GC properties towards the osteoblast phenotype. The present study, therefore, suggests that GCs may become an excellent starting material in obtaining stable osteoblast cultures. GCs differentiated towards osteoblasts may be used in regenerative and reconstructive medicine in the future.
Figures

References
-
- Rybska M, Knap S, Jankowski M, Jeseta M, Bukowska D, Antosik P, Nowicki M, Zabel M, Kempisty B, Jaśkowski JM. Characteristic of factors influencing the proper course of folliculogenesis in mammals. Med J Cell Biol. 2018;6:33–38. doi: 10.2478/acb-2018-0006. - DOI
-
- Kranc W, Jankowski M, Budna J, Celichowski P, Khozmi R, Bryja A, Borys S, Dyszkiewicz-Konwińska M, Jeseta M, Magas M, et al. Amino acids metabolism and degradation is regulated during porcine oviductal epithelial cells (OECs) primary culture in vitro-signaling pathway activation approach. Med J Cell Biol. 2018;6:18–26. doi: 10.2478/acb-2018-0004. - DOI
-
- Rybska M, Knap S, Jankowski M, Jeseta M, Bukowska D, Antosik P, Nowicki M, Zabel M, Kempisty B, Jaśkowski JM. Cytoplasmic and nuclear maturation of oocytes in mammals-living in the shadow of cells developmental capability. Med J Cell Biol. 2018;1:13–17. doi: 10.2478/acb-2018-0003. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

