Dissecting the Energetics of Intrinsically Disordered Proteins via a Hybrid Experimental and Computational Approach
- PMID: 31702919
- PMCID: PMC7291390
- DOI: 10.1021/acs.jpcb.9b08323
Dissecting the Energetics of Intrinsically Disordered Proteins via a Hybrid Experimental and Computational Approach
Abstract
Intrinsically disordered proteins (IDPs) play important roles in biology, but little is known about the energetics of their inter-residue interactions. Methods that have been successfully applied to analyze the energetics of globular proteins are not applicable to the fluctuating partially ordered ensembles populated by IDPs. A combined computational experimental strategy is introduced for analyzing the energetic role of individual residues in the free state of IDPs. The approach combines experimental measurements of the binding of wild-type and mutant IDPs to their partners with alchemical free energy calculations of the structured complexes. These data allow quantitative information to be deduced about the free state via a thermodynamic cycle. The approach is validated by the analysis of the effects of mutations upon the binding free energy of the ovomucoid inhibitor third binding domain to its partners and is applied to the C-terminal domain of the measles virus nucleoprotein, a 125-residue IDP involved in the RNA transcription and replication of measles virus. The analysis reveals significant inter-residue interactions in the unbound IDP and suggests a biological role for them. The work demonstrates that advances in force fields and computational hardware have now led to the point where it is possible to develop methods, which integrate experimental and computational techniques to reveal insights that cannot be studied using either technique alone.
Figures


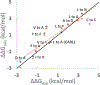

References
-
- Romero P; Obradovic Z; Li X; Garner EC; Brown CJ; Dunker AK, Sequence complexity of disordered protein. Proteins 2001, 42 (1), 38–48. - PubMed
-
- Oldfield CJ; Dunker AK, Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 2014, 83, 553–584. - PubMed
-
- Dyson HJ; Wright PE, Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005, 6 (3), 197–208. - PubMed

