Functional categorization of de novo transcriptome assembly of Vanilla planifolia Jacks. potentially points to a translational regulation during early stages of infection by Fusarium oxysporum f. sp. vanillae
- PMID: 31703622
- PMCID: PMC6839141
- DOI: 10.1186/s12864-019-6229-5
Functional categorization of de novo transcriptome assembly of Vanilla planifolia Jacks. potentially points to a translational regulation during early stages of infection by Fusarium oxysporum f. sp. vanillae
Abstract
Background: Upon exposure to unfavorable environmental conditions, plants need to respond quickly to maintain their homeostasis. For instance, physiological, biochemical and transcriptional changes occur during plant-pathogen interaction. In the case of Vanilla planifolia Jacks., a worldwide economically important crop, it is susceptible to Fusarium oxysporum f. sp. vanillae (Fov). This pathogen causes root and stem rot (RSR) in vanilla plants that lead to plant death. To investigate how vanilla plants, respond at the transcriptional level upon infection with Fov, here we employed the RNA-Seq approach to analyze the dynamics of whole-transcriptome changes during two-time frames of the infection.
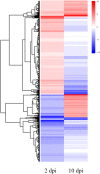
Results: Analysis of global gene expression profiles upon infection by Fov indicated that the major transcriptional change occurred at 2 days post-inoculation (dpi), in comparison to 10 dpi. Briefly, the RNA-Seq analysis carried out in roots found that 3420 and 839 differentially expressed genes (DEGs) were detected at 2 and 10 dpi, respectively, as compared to the control. In the case of DEGs at 2 dpi, 1563 genes were found to be up-regulated, whereas 1857 genes were down-regulated. Moreover, functional categorization of DEGs at 2 dpi indicated that up-regulated genes are mainly associated to translation, whereas down-regulated genes are involved in cell wall remodeling. Among the translational-related transcripts, ribosomal proteins (RPs) were found increased their expression exclusively at 2 dpi.
Conclusions: The screening of transcriptional changes of V. planifolia Jacks upon infection by Fov provides insights into the plant molecular response, particularly at early stages of infection. The accumulation of translational-related transcripts at early stages of infection potentially points to a transcriptional reprogramming coupled with a translational regulation in vanilla plants upon infection by Fov. Altogether, the results presented here highlight potential molecular players that might be further studied to improve Fov-induced resistance in vanilla plants.
Keywords: Biological defense; Biotic stress; Ribosomal proteins; Transcriptional reprogramming; Translational regulation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

