Structures of SF3b1 reveal a dynamic Achilles heel of spliceosome assembly: Implications for cancer-associated abnormalities and drug discovery
- PMID: 31707043
- PMCID: PMC6880240
- DOI: 10.1016/j.bbagrm.2019.194440
Structures of SF3b1 reveal a dynamic Achilles heel of spliceosome assembly: Implications for cancer-associated abnormalities and drug discovery
Abstract
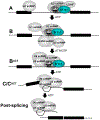
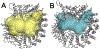
The pre-mRNA splicing factor SF3b1 exhibits recurrent mutations among hematologic malignancies and cancers, and consequently is a major therapeutic target of clinically-advanced spliceosome inhibitors. In this review, we highlight and rigorously analyze emerging views of SF3b1 conformational transitions, including the human SF3b particle either in isolation or bound to spliceosome inhibitors, and human or yeast spliceosome assemblies. Among spliceosome states characterized to date, an SF3b1 α-helical superhelix significantly closes to surround a U2 small nuclear RNA duplex with the pre-mRNA branch point sequence. The SF3b1 torus is locally unwound at an active site adenosine, whereas protein cofactors appear to stabilize overall closure in the spliceosome. Network analyses demonstrates that the natural SF3b1 dynamics mimic its conformational change in the spliceosome, raising the possibility of conformational selection underpinning spliceosome assembly. These dynamic SF3b1 conformations have consequences for gatekeeping of spliceosome assembly and therapeutic targeting of its cancer-associated dysfunction.
Keywords: HEAT repeat; Hsh155; Myelodysplastic syndrome; Pre-mRNA splicing, cancer; Ribonucleoprotein structure.
Copyright © 2019 Elsevier B.V. All rights reserved.
Figures
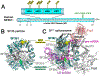

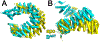


Similar articles
-
SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing.Genes Dev. 2016 Dec 15;30(24):2710-2723. doi: 10.1101/gad.291872.116. Genes Dev. 2016. PMID: 28087715 Free PMC article.
-
The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.J Biol Chem. 2019 Feb 22;294(8):2892-2902. doi: 10.1074/jbc.RA118.006764. Epub 2018 Dec 19. J Biol Chem. 2019. PMID: 30567737 Free PMC article.
-
Impact of cancer-associated mutations in Hsh155/SF3b1 HEAT repeats 9-12 on pre-mRNA splicing in Saccharomyces cerevisiae.PLoS One. 2020 Apr 22;15(4):e0229315. doi: 10.1371/journal.pone.0229315. eCollection 2020. PLoS One. 2020. PMID: 32320410 Free PMC article.
-
The SF3b Complex is an Integral Component of the Spliceosome and Targeted by Natural Product-Based Inhibitors.Subcell Biochem. 2021;96:409-432. doi: 10.1007/978-3-030-58971-4_12. Subcell Biochem. 2021. PMID: 33252738 Review.
-
Splicing Factor Mutations in Myelodysplasias: Insights from Spliceosome Structures.Trends Genet. 2017 May;33(5):336-348. doi: 10.1016/j.tig.2017.03.001. Epub 2017 Mar 31. Trends Genet. 2017. PMID: 28372848 Free PMC article. Review.
Cited by
-
The SF3b complex: splicing and beyond.Cell Mol Life Sci. 2020 Sep;77(18):3583-3595. doi: 10.1007/s00018-020-03493-z. Epub 2020 Mar 5. Cell Mol Life Sci. 2020. PMID: 32140746 Free PMC article. Review.
-
Variations of intronic branchpoint motif: identification and functional implications in splicing and disease.Commun Biol. 2023 Nov 10;6(1):1142. doi: 10.1038/s42003-023-05513-7. Commun Biol. 2023. PMID: 37949953 Free PMC article. Review.
-
Systematic Profiling of mRNA Splicing Reveals the Prognostic Predictor and Potential Therapeutic Target for Glioblastoma Multiforme.J Oncol. 2021 Jul 5;2021:4664955. doi: 10.1155/2021/4664955. eCollection 2021. J Oncol. 2021. PMID: 34326872 Free PMC article.
-
[Screening of drugs that selectively inhibit uveal melanoma cells with SF3B1 mutations].Nan Fang Yi Ke Da Xue Xue Bao. 2021 Dec 20;41(12):1835-1842. doi: 10.12122/j.issn.1673-4254.2021.12.12. Nan Fang Yi Ke Da Xue Xue Bao. 2021. PMID: 35012916 Free PMC article. Chinese.
-
Small molecules modulating RNA splicing: a review of targets and future perspectives.RSC Med Chem. 2024 Jan 11;15(4):1109-1126. doi: 10.1039/d3md00685a. eCollection 2024 Apr 24. RSC Med Chem. 2024. PMID: 38665842 Free PMC article. Review.
References
-
- Agrawal AA, Yu L, Smith PG, Buonamici S, Targeting splicing abnormalities in cancer, Curr Opin Genet Dev, 48 (2018) 67–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

