Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells
- PMID: 31708126
- PMCID: PMC6904908
- DOI: 10.1016/j.cell.2019.10.015
Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells
Abstract
The colonic epithelium can undergo multiple rounds of damage and repair, often in response to excessive inflammation. The responsive stem cell that mediates this process is unclear, in part because of a lack of in vitro models that recapitulate key epithelial changes that occur in vivo during damage and repair. Here, we identify a Hopx+ colitis-associated regenerative stem cell (CARSC) population that functionally contributes to mucosal repair in mouse models of colitis. Hopx+ CARSCs, enriched for fetal-like markers, transiently arose from hypertrophic crypts known to facilitate regeneration. Importantly, we established a long-term, self-organizing two-dimensional (2D) epithelial monolayer system to model the regenerative properties and responses of Hopx+ CARSCs. This system can reenact the "homeostasis-injury-regeneration" cycles of epithelial alterations that occur in vivo. Using this system, we found that hypoxia and endoplasmic reticulum stress, insults commonly present in inflammatory bowel diseases, mediated the cyclic switch of cellular status in this process.
Keywords: HopX; Lgr5; Transwell; air-liquid interface; colitis; colon; hypoxia; intestine; stem cell; unfolded protein response.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
T.S.S. has served on an advisory board for Boehringer Ingelheim.
Figures

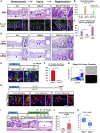


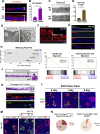




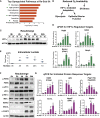

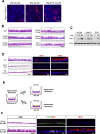
Comment in
-
A new model of intestinal epithelial regeneration: could patients benefit?Nat Rev Gastroenterol Hepatol. 2020 Mar;17(3):137-138. doi: 10.1038/s41575-020-0267-y. Nat Rev Gastroenterol Hepatol. 2020. PMID: 31959899 No abstract available.
References
-
- Bader M., Muse W., Ballou D.P., Gassner C., Bardwell J.C. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. - PubMed
-
- Baert F., Moortgat L., Van Assche G., Caenepeel P., Vergauwe P., De Vos M., Stokkers P., Hommes D., Rutgeerts P., Vermeire S., D’Haens G., Belgian Inflammatory Bowel Disease Research Group. North-Holland Gut Club Mucosal healing predicts sustained clinical remission in patients with early-stage Crohn’s disease. Gastroenterology. 2010;138:463–468. - PubMed
-
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., NIDDK IBD Genetics Consortium. Belgian-French IBD Consortium. Wellcome Trust Case Control Consortium Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

