Wnt Inhibition Facilitates RNA-Mediated Reprogramming of Human Somatic Cells to Naive Pluripotency
- PMID: 31708477
- PMCID: PMC6915845
- DOI: 10.1016/j.stemcr.2019.10.009
Wnt Inhibition Facilitates RNA-Mediated Reprogramming of Human Somatic Cells to Naive Pluripotency
Abstract
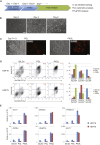
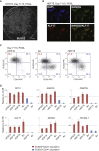
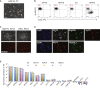
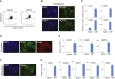
In contrast to conventional human pluripotent stem cells (hPSCs) that are related to post-implantation embryo stages, naive hPSCs exhibit features of pre-implantation epiblast. Naive hPSCs are established by resetting conventional hPSCs, or are derived from dissociated embryo inner cell masses. Here we investigate conditions for transgene-free reprogramming of human somatic cells to naive pluripotency. We find that Wnt inhibition promotes RNA-mediated induction of naive pluripotency. We demonstrate application to independent human fibroblast cultures and endothelial progenitor cells. We show that induced naive hPSCs can be clonally expanded with a diploid karyotype and undergo somatic lineage differentiation following formative transition. Induced naive hPSC lines exhibit distinctive surface marker, transcriptome, and methylome properties of naive epiblast identity. This system for efficient, facile, and reliable induction of transgene-free naive hPSCs offers a robust platform, both for delineation of human reprogramming trajectories and for evaluating the attributes of isogenic naive versus conventional hPSCs.
Keywords: RNA-mediated reprogramming; Wnt signaling; human pluripotent stem cells; human pre-implantation epiblast; induced pluripotent stem cells; molecular reprogramming; naive pluripotency.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., Bousson F., Zidouni Y., Mursch C., Moncuquet P. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015;33:962–969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

