Deciphering the Functioning of Microbial Communities: Shedding Light on the Critical Steps in Metaproteomics
- PMID: 31708885
- PMCID: PMC6821674
- DOI: 10.3389/fmicb.2019.02395
Deciphering the Functioning of Microbial Communities: Shedding Light on the Critical Steps in Metaproteomics
Abstract
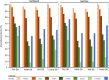
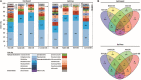
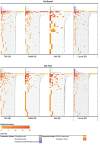
Unraveling the complex structure and functioning of microbial communities is essential to accurately predict the impact of perturbations and/or environmental changes. From all molecular tools available today to resolve the dynamics of microbial communities, metaproteomics stands out, allowing the establishment of phenotype-genotype linkages. Despite its rapid development, this technology has faced many technical challenges that still hamper its potential power. How to maximize the number of protein identification, improve quality of protein annotation, and provide reliable ecological interpretation are questions of immediate urgency. In our study, we used a robust metaproteomic workflow combining two protein fractionation approaches (gel-based versus gel-free) and four protein search databases derived from the same metagenome to analyze the same seawater sample. The resulting eight metaproteomes provided different outcomes in terms of (i) total protein numbers, (ii) taxonomic structures, and (iii) protein functions. The characterization and/or representativeness of numerous proteins from ecologically relevant taxa such as Pelagibacterales, Rhodobacterales, and Synechococcales, as well as crucial environmental processes, such as nutrient uptake, nitrogen assimilation, light harvesting, and oxidative stress response, were found to be particularly affected by the methodology. Our results provide clear evidences that the use of different protein search databases significantly alters the biological conclusions in both gel-free and gel-based approaches. Our findings emphasize the importance of diversifying the experimental workflow for a comprehensive metaproteomic study.
Keywords: bioinformatics; mass spectrometry; metagenomics; metaproteomics; microbial ecology.
Copyright © 2019 Géron, Werner, Wattiez, Lebaron and Matallana-Surget.
Figures


References
-
- Absciex (2014). Understanding the Pro GroupTM Algorithm. Available at: https://sciex.com/Documents/manuals/proteinPilot-ProGroup-Algorithm.pdf (accessed June 12, 2019).
-
- Button D. K., Robertson B. (2000). Effect of nutrient kinetics and cytoarchitecture on bacterioplankter size. Limnol. Oceanogr. 45 499–505. 10.4319/lo.2000.45.2.0499 - DOI
LinkOut - more resources
Full Text Sources

