A Bispecific Antibody to Link a TRAIL-Based Antitumor Approach to Immunotherapy
- PMID: 31708930
- PMCID: PMC6823250
- DOI: 10.3389/fimmu.2019.02514
A Bispecific Antibody to Link a TRAIL-Based Antitumor Approach to Immunotherapy
Abstract
T-cell-based immunotherapy strategies have profoundly improved the clinical management of several solid tumors and hematological malignancies. A recently developed and promising immunotherapy approach is to redirect polyclonal MHC-unrestricted T lymphocytes toward cancer cells by bispecific antibodies (bsAbs) that engage the CD3 complex and a tumor-associated antigen (TAA). The TNF-related apoptosis-inducing ligand receptor 2 (TRAIL-R2) is an attractive immunotherapy target, frequently expressed by neoplastic cells, that we decided to exploit as a TAA. We found that a TRAIL-R2xCD3 bsAb efficiently activates T cells and specifically redirect their cytotoxicity against cancer cells of different origins in vitro, thereby demonstrating its potential as a pan-carcinoma reagent. Moreover, to mimic in vivo conditions, we assessed its ability to retarget T-cell activity in an ex vivo model of ovarian cancer patients' ascitic fluids containing both effector and target cells-albeit with a suboptimal effector-to-target ratio-with remarkable results.
Keywords: T-cell retargeting; TRAIL-R2; bispecific antibody; immunotherapy; malignant ascites.
Copyright © 2019 Satta, Grazia, Caroli, Frigerio, Di Nicola, Raspagliesi, Mezzanzanica, Zaffaroni, Gianni, Anichini and Figini.
Figures
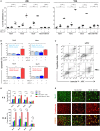


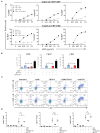
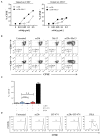
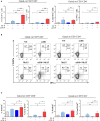
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

