Development of Unconventional T Cells Controlled by MicroRNA
- PMID: 31708931
- PMCID: PMC6820353
- DOI: 10.3389/fimmu.2019.02520
Development of Unconventional T Cells Controlled by MicroRNA
Abstract
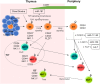
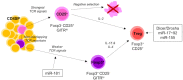
Post-transcriptional gene regulation through microRNA (miRNA) has emerged as a major control mechanism of multiple biological processes, including development and function of T cells. T cells are vital components of the immune system, with conventional T cells playing a central role in adaptive immunity and unconventional T cells having additional functions reminiscent of both innate and adaptive immunity, such as involvement in stress responses and tissue homeostasis. Unconventional T cells encompass cells expressing semi-invariant T cell receptors (TCRs), such as invariant Natural Killer T (iNKT) and Mucosal-Associated Invariant T (MAIT) cells. Additionally, some T cells with diverse TCR repertoires, including γδT cells, intraepithelial lymphocytes (IEL) and regulatory T (Treg) cells, share some functional and/or developmental features with their semi-invariant unconventional counterparts. Unconventional T cells are particularly sensitive to disruption of miRNA function, both globally and on the individual miRNA level. Here, we review the role of miRNA in the development and function of unconventional T cells from an iNKT-centric point of view. The function of single miRNAs can provide important insights into shared and individual pathways for the formation of different unconventional T cell subsets.
Keywords: MAIT cell; NKT cell; T cell; Treg cell; miR-181; microRNA; thymus; γδT cell.
Copyright © 2019 Winter and Krueger.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

