New Vocabulary for Bacterial Communication
- PMID: 31709676
- PMCID: PMC7154725
- DOI: 10.1002/cbic.201900580
New Vocabulary for Bacterial Communication
Abstract
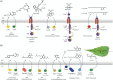
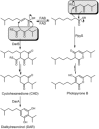
Quorum sensing (QS) is widely accepted as a procedure that bacteria use to converse. However, prevailing thinking places acyl homoserine lactones (AHLs) at the forefront of this communication pathway in Gram-negative bacteria. With the advent of high-throughput genomics and the subsequent influx of bacterial genomes, bioinformatics analysis has determined that the genes encoding AHL biosynthesis, originally discovered to be indispensable for QS (LuxI-like proteins and homologues), are often absent in QS-capable bacteria. Instead, the sensing protein (LuxR-like proteins) is present with an apparent inability to produce any outgoing AHL signal. Recently, several signals for these LuxR solos have been identified. Herein, advances in the field of QS are discussed, with a particular focus on recent research in the field of bacterial cell-cell communication.
Keywords: bacterial communication; biosynthesis; gene sequencing; quorum quenching; quorum sensing.
© 2019 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Conflict of interest statement
Figures

References
-
- Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P., Science 1998, 280, 295–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

