"Trim"ming PolyQ proteins with engineered PML
- PMID: 31710088
- PMCID: PMC6952540
- DOI: 10.1002/bit.27220
"Trim"ming PolyQ proteins with engineered PML
Abstract
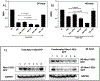
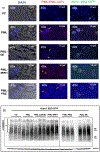
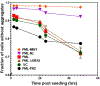
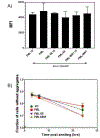
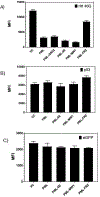
Protein abnormalities are the major cause of neurodegenerative diseases such as spinocerebellar ataxia (SCA). Protein misfolding and impaired degradation leads to the build-up of protein aggregates inside the cell, which may further cause cellular degeneration. Reducing levels of either the soluble misfolded form of the protein or its precipitated aggregate, even marginally, could significantly improve cellular health. Despite numerous pre-existing strategies to target these protein aggregates, there is considerable room to improve their specificity and efficiency. In this study, we demonstrated the enhanced intracellular degradation of both monomers and aggregates of mutant ataxin1 (Atxn1 82Q) by engineering an E3 ubiquitin ligase enzyme, promyelocytic leukemia protein (PML). Specifically, we showed enhanced degradation of both soluble and aggregated Atxn1 82Q in mammalian cells by targeting this protein using PML fused to single chain variable fragments (scFvs) specific for monomers and aggregates of the target protein. The ability to solubilize Atxn1 82Q aggregates was due to the PML-mediated enhanced SUMOylation of the target protein. This ability to reduce the intracellular levels of both misfolded forms of Atxn1 82Q may not only be useful for treating SCA, but also applicable for the treatment of other PolyQ disorders.
Keywords: PML; aggregate; degradation; monomer; spinocerebellar ataxia.
© 2019 Wiley Periodicals, Inc.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

