Developing a Reproducible Microbiome Data Analysis Pipeline Using the Amazon Web Services Cloud for a Cancer Research Group: Proof-of-Concept Study
- PMID: 31710301
- PMCID: PMC6913755
- DOI: 10.2196/14667
Developing a Reproducible Microbiome Data Analysis Pipeline Using the Amazon Web Services Cloud for a Cancer Research Group: Proof-of-Concept Study
Abstract
Background: Cloud computing for microbiome data sets can significantly increase working efficiencies and expedite the translation of research findings into clinical practice. The Amazon Web Services (AWS) cloud provides an invaluable option for microbiome data storage, computation, and analysis.
Objective: The goals of this study were to develop a microbiome data analysis pipeline by using AWS cloud and to conduct a proof-of-concept test for microbiome data storage, processing, and analysis.
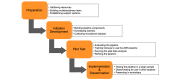
Methods: A multidisciplinary team was formed to develop and test a reproducible microbiome data analysis pipeline with multiple AWS cloud services that could be used for storage, computation, and data analysis. The microbiome data analysis pipeline developed in AWS was tested by using two data sets: 19 vaginal microbiome samples and 50 gut microbiome samples.
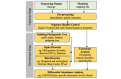
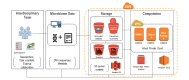
Results: Using AWS features, we developed a microbiome data analysis pipeline that included Amazon Simple Storage Service for microbiome sequence storage, Linux Elastic Compute Cloud (EC2) instances (ie, servers) for data computation and analysis, and security keys to create and manage the use of encryption for the pipeline. Bioinformatics and statistical tools (ie, Quantitative Insights Into Microbial Ecology 2 and RStudio) were installed within the Linux EC2 instances to run microbiome statistical analysis. The microbiome data analysis pipeline was performed through command-line interfaces within the Linux operating system or in the Mac operating system. Using this new pipeline, we were able to successfully process and analyze 50 gut microbiome samples within 4 hours at a very low cost (a c4.4xlarge EC2 instance costs $0.80 per hour). Gut microbiome findings regarding diversity, taxonomy, and abundance analyses were easily shared within our research team.
Conclusions: Building a microbiome data analysis pipeline with AWS cloud is feasible. This pipeline is highly reliable, computationally powerful, and cost effective. Our AWS-based microbiome analysis pipeline provides an efficient tool to conduct microbiome data analysis.
Keywords: Amazon Web Services; cloud computation; microbiome; pipeline; sequence analysis.
©Jinbing Bai, Ileen Jhaney, Jessica Wells. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 11.11.2019.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
References
-
- Prosperi M, Min JS, Bian J, Modave F. Big data hurdles in precision medicine and precision public health. BMC Med Inform Decis Mak. 2018 Dec 29;18(1):139. doi: 10.1186/s12911-018-0719-2. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-0... - DOI - DOI - PMC - PubMed
-
- Agarwal V, Zhang L, Zhu J, Fang S, Cheng T, Hong C, Shah NH. Impact of Predicting Health Care Utilization Via Web Search Behavior: A Data-Driven Analysis. J Med Internet Res. 2016 Sep 21;18(9):e251. doi: 10.2196/jmir.6240. https://www.jmir.org/2016/9/e251/ - DOI - PMC - PubMed
-
- Ursell Luke K, Metcalf Jessica L, Parfrey Laura Wegener, Knight Rob. Defining the human microbiome. Nutr Rev. 2012 Aug;70 Suppl 1:S38–44. doi: 10.1111/j.1753-4887.2012.00493.x. http://europepmc.org/abstract/MED/22861806 - DOI - PMC - PubMed
-
- Knight R. Follow Your Gut: The Enormous Impact Of Tiny Microbes (TED Books) Simon & Schuster / Ted; 2019.
-
- Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016 Jan 28;164(3):337–40. doi: 10.1016/j.cell.2016.01.013. https://linkinghub.elsevier.com/retrieve/pii/S0092-8674(16)00053-2 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

