Adaptation in structured populations and fuzzy boundaries between hard and soft sweeps
- PMID: 31710623
- PMCID: PMC6872172
- DOI: 10.1371/journal.pcbi.1007426
Adaptation in structured populations and fuzzy boundaries between hard and soft sweeps
Abstract
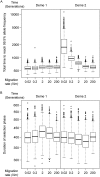
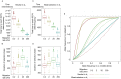
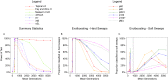
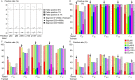
Selective sweeps, the genetic footprint of positive selection, have been extensively studied in the past decades, with dozens of methods developed to identify swept regions. However, these methods suffer from both false positive and false negative reports, and the candidates identified with different methods are often inconsistent with each other. We propose that a biological cause of this problem can be population subdivision, and a technical cause can be incomplete, or inaccurate, modeling of the dynamic process associated with sweeps. Here we used simulations to show how these effects interact and potentially cause bias. In particular, we show that sweeps maybe misclassified as either hard or soft, when the true time stage of a sweep and that implied, or pre-supposed, by the model do not match. We call this "temporal misclassification". Similarly, "spatial misclassification (softening)" can occur when hard sweeps, which are imported by migration into a new subpopulation, are falsely identified as soft. This can easily happen in case of local adaptation, i.e. when the sweeping allele is not under positive selection in the new subpopulation, and the underlying model assumes panmixis instead of substructure. The claim that most sweeps in the evolutionary history of humans were soft, may have to be reconsidered in the light of these findings.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


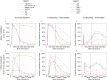
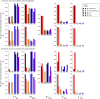
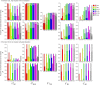

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

