The Dundee Resource for Sequence Analysis and Structure Prediction
- PMID: 31710725
- PMCID: PMC6933851
- DOI: 10.1002/pro.3783
The Dundee Resource for Sequence Analysis and Structure Prediction
Abstract
The Dundee Resource for Sequence Analysis and Structure Prediction (DRSASP; http://www.compbio.dundee.ac.uk/drsasp.html) is a collection of web services provided by the Barton Group at the University of Dundee. DRSASP's flagship services are the JPred4 webserver for secondary structure and solvent accessibility prediction and the JABAWS 2.2 webserver for multiple sequence alignment, disorder prediction, amino acid conservation calculations, and specificity-determining site prediction. DRSASP resources are available through conventional web interfaces and APIs but are also integrated into the Jalview sequence analysis workbench, which enables the composition of multitool interactive workflows. Other existing Barton Group tools are being brought under the banner of DRSASP, including NoD (Nucleolar localization sequence detector) and 14-3-3-Pred. New resources are being developed that enable the analysis of population genetic data in evolutionary and 3D structural contexts. Existing resources are actively developed to exploit new technologies and maintain parity with evolving web standards. DRSASP provides substantial computational resources for public use, and since 2016 DRSASP services have completed over 1.5 million jobs.
© 2019 The Authors. Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Figures


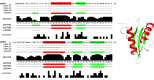

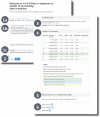
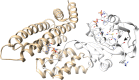

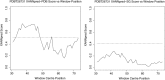

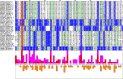

References
Publication types
MeSH terms
Substances
Grants and funding
- BB/J019364/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 101651/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- BB/G022682/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R014752/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

