The Evening Complex Establishes Repressive Chromatin Domains Via H2A.Z Deposition
- PMID: 31712305
- PMCID: PMC6945876
- DOI: 10.1104/pp.19.00881
The Evening Complex Establishes Repressive Chromatin Domains Via H2A.Z Deposition
Abstract
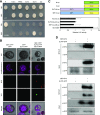
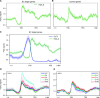
The Evening Complex (EC) is a core component of the Arabidopsis (Arabidopsis thaliana) circadian clock, which represses target gene expression at the end of the day and integrates temperature information to coordinate environmental and endogenous signals. Here we show that the EC induces repressive chromatin structure to regulate the evening transcriptome. The EC component ELF3 directly interacts with a protein from the SWI2/SNF2-RELATED (SWR1) complex to control deposition of H2A.Z-nucleosomes at the EC target genes. SWR1 components display circadian oscillation in gene expression with a peak at dusk. In turn, SWR1 is required for the circadian clockwork, as defects in SWR1 activity alter morning-expressed genes. The EC-SWR1 complex binds to the loci of the core clock genes PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9 and catalyzes deposition of nucleosomes containing the histone variant H2A.Z coincident with the repression of these genes at dusk. This provides a mechanism by which the circadian clock temporally establishes repressive chromatin domains to shape oscillatory gene expression around dusk.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 - PubMed
-
- Carré IA, Kim JY (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53: 1551–1557 - PubMed

