Accelerated evolution of oligodendrocytes in the human brain
- PMID: 31712436
- PMCID: PMC6883816
- DOI: 10.1073/pnas.1907982116
Accelerated evolution of oligodendrocytes in the human brain
Abstract
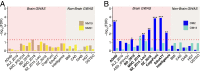
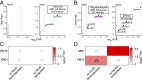
Recent discussions of human brain evolution have largely focused on increased neuron numbers and changes in their connectivity and expression. However, it is increasingly appreciated that oligodendrocytes play important roles in cognitive function and disease. Whether both cell types follow similar or distinctive evolutionary trajectories is not known. We examined the transcriptomes of neurons and oligodendrocytes in the frontal cortex of humans, chimpanzees, and rhesus macaques. We identified human-specific trajectories of gene expression in neurons and oligodendrocytes and show that both cell types exhibit human-specific up-regulation. Moreover, oligodendrocytes have undergone more pronounced accelerated gene expression evolution in the human lineage compared to neurons. We highlighted human-specific coexpression networks with specific functions. Our data suggest that oligodendrocyte human-specific networks are enriched for alternative splicing and transcriptional regulation. Oligodendrocyte networks are also enriched for variants associated with schizophrenia and other neuropsychiatric disorders. Such enrichments were not found in neuronal networks. These results offer a glimpse into the molecular mechanisms of oligodendrocytes during evolution and how such mechanisms are associated with neuropsychiatric disorders.
Keywords: brain evolution; cell-type expression; comparative primate genomics; neurogenomics.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Preuss T. M., “The human brain: Evolution and distinctive features” in On Human Nature, Tibayrenc M., Ayala F. J., Eds. (Academic Press, San Diego, 2017), chap. 8, pp. 125–149.
-
- Khaitovich P., Enard W., Lachmann M., Pääbo S., Evolution of primate gene expression. Nat. Rev. Genet. 7, 693–702 (2006). - PubMed
-
- King M. C., Wilson A. C., Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

