An Emerging Clone, Klebsiellapneumoniae Carbapenemase 2-Producing K. pneumoniae Sequence Type 16, Associated With High Mortality Rates in a CC258-Endemic Setting
- PMID: 31712802
- PMCID: PMC7583420
- DOI: 10.1093/cid/ciz1095
An Emerging Clone, Klebsiellapneumoniae Carbapenemase 2-Producing K. pneumoniae Sequence Type 16, Associated With High Mortality Rates in a CC258-Endemic Setting
Abstract
Background: Carbapenemase-producing Klebsiella pneumoniae has become a global priority, not least in low- and middle-income countries. Here, we report the emergence and clinical impact of a novel Klebsiella pneumoniae carbapenemase-producing K. pneumoniae (KPC-KP) sequence type (ST) 16 clone in a clonal complex (CC) 258-endemic setting.
Methods: In a teaching Brazilian hospital, a retrospective cohort of adult KPC-KP bloodstream infection (BSI) cases (January 2014 to December 2016) was established to study the molecular epidemiology and its impact on outcome (30-day all-cause mortality). KPC-KP isolates underwent multilocus sequence typing. Survival analysis between ST/CC groups and risk factors for fatal outcome (logistic regression) were evaluated. Representative isolates underwent whole-genome sequencing and had their virulence tested in a Galleria larvae model.
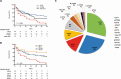
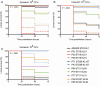
Results: One hundred sixty-five unique KPC-KP BSI cases were identified. CC258 was predominant (66%), followed by ST16 (12%). The overall 30-day mortality rate was 60%; in contrast, 95% of ST16 cases were fatal. Patients' severity scores were high and baseline clinical variables were not statistically different across STs. In multivariate analysis, ST16 (odds ratio [OR], 21.4; 95% confidence interval [CI], 2.3-202.8; P = .008) and septic shock (OR, 11.9; 95% CI, 4.2-34.1; P < .001) were independent risk factors for fatal outcome. The ST16 clone carried up to 14 resistance genes, including blaKPC-2 in an IncFIBpQIL plasmid, KL51 capsule, and yersiniabactin virulence determinants. The ST16 clone was highly pathogenic in the larvae model.
Conclusions: Mortality rates were high in this KPC-KP BSI cohort, where CC258 is endemic. An emerging ST16 clone was associated with high mortality. Our results suggest that even in endemic settings, highly virulent clones can rapidly emerge demanding constant monitoring.
Keywords: Klebsiella pneumoniae; CC258; KPC; bloodstream infections; carbapenem-resistant Enterobacteriaceae.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures

References
-
- Centers for Diseases Control and Prevention. Carbapenem-resistant Enterobacteriaceae (CRE). Available at: https://www.cdc.gov/hai/organisms/cre/index.html. Accessed 18 January 2019.
-
- World Health Organization. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. Available at: http://www.who.int/infection-prevention/publications/guidelines-cre/en/. Accessed 18 January 2019. - PubMed
-
- Public Health England. Carbapenem resistance: guidance, data and analysis. Available at: https://www.gov.uk/government/collections/carbapenem-resistance-guidance.... Accessed 18 January 2019.
-
- Antochevis LC, Magagnin CM, Nunes AG, et al. KPC-producing Klebsiella pneumoniae bloodstream isolates from Brazilian hospitals: what (still) remains active? J Glob Antimicrob Resist 2018; 15:173–7. - PubMed

