Dynamics of the DEAD-box ATPase Prp5 RecA-like domains provide a conformational switch during spliceosome assembly
- PMID: 31712821
- PMCID: PMC6846040
- DOI: 10.1093/nar/gkz765
Dynamics of the DEAD-box ATPase Prp5 RecA-like domains provide a conformational switch during spliceosome assembly
Abstract
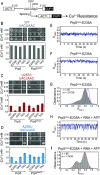
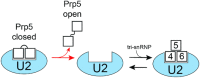
The DEAD-box family of proteins are ATP-dependent, RNA-binding proteins implicated in many aspects of RNA metabolism. Pre-mRNA splicing in eukaryotes requires three DEAD-box ATPases (Prp5, Prp28 and Sub2), the molecular mechanisms of which are poorly understood. Here, we use single molecule FRET (smFRET) to study the conformational dynamics of yeast Prp5. Prp5 is essential for stable association of the U2 snRNP with the intron branch site (BS) sequence during spliceosome assembly. Our data show that the Prp5 RecA-like domains undergo a large conformational rearrangement only in response to binding of both ATP and RNA. Mutations in Prp5 impact the fidelity of BS recognition and change the conformational dynamics of the RecA-like domains. We propose that BS recognition during spliceosome assembly involves a set of coordinated conformational switches among U2 snRNP components. Spontaneous toggling of Prp5 into a stable, open conformation may be important for its release from U2 and to prevent competition between Prp5 re-binding and subsequent steps in spliceosome assembly.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Rudolph M.G., Klostermeier D.. When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol. Chem. 2015; 396:849–865. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

