New developments on the Encyclopedia of DNA Elements (ENCODE) data portal
- PMID: 31713622
- PMCID: PMC7061942
- DOI: 10.1093/nar/gkz1062
New developments on the Encyclopedia of DNA Elements (ENCODE) data portal
Abstract
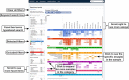
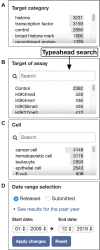
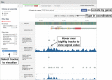
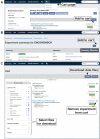
The Encyclopedia of DNA Elements (ENCODE) is an ongoing collaborative research project aimed at identifying all the functional elements in the human and mouse genomes. Data generated by the ENCODE consortium are freely accessible at the ENCODE portal (https://www.encodeproject.org/), which is developed and maintained by the ENCODE Data Coordinating Center (DCC). Since the initial portal release in 2013, the ENCODE DCC has updated the portal to make ENCODE data more findable, accessible, interoperable and reusable. Here, we report on recent updates, including new ENCODE data and assays, ENCODE uniform data processing pipelines, new visualization tools, a dataset cart feature, unrestricted public access to ENCODE data on the cloud (Amazon Web Services open data registry, https://registry.opendata.aws/encode-project/) and more comprehensive tutorials and documentation.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. - PubMed
-
- The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004; 306:636–640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

