Lipid lowering and Alzheimer disease risk: A mendelian randomization study
- PMID: 31714636
- PMCID: PMC6944510
- DOI: 10.1002/ana.25642
Lipid lowering and Alzheimer disease risk: A mendelian randomization study
Abstract
Objective: To examine whether genetic variation affecting the expression or function of lipid-lowering drug targets is associated with Alzheimer disease (AD) risk, to evaluate the potential impact of long-term exposure to corresponding therapeutics.
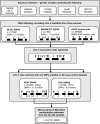
Methods: We conducted Mendelian randomization analyses using variants in genes that encode the protein targets of several approved lipid-lowering drug classes: HMGCR (encoding the target for statins), PCSK9 (encoding the target for PCSK9 inhibitors, eg, evolocumab and alirocumab), NPC1L1 (encoding the target for ezetimibe), and APOB (encoding the target of mipomersen). Variants were weighted by associations with low-density lipoprotein cholesterol (LDL-C) using data from lipid genetics consortia (n up to 295,826). We meta-analyzed Mendelian randomization estimates for regional variants weighted by LDL-C on AD risk from 2 large samples (total n = 24,718 cases, 56,685 controls).
Results: Models for HMGCR, APOB, and NPC1L1 did not suggest that the use of related lipid-lowering drug classes would affect AD risk. In contrast, genetically instrumented exposure to PCSK9 inhibitors was predicted to increase AD risk in both of the AD samples (combined odds ratio per standard deviation lower LDL-C inducible by the drug target = 1.45, 95% confidence interval = 1.23-1.69). This risk increase was opposite to, although more modest than, the degree of protection from coronary artery disease predicted by these same methods for PCSK9 inhibition.
Interpretation: We did not identify genetic support for the repurposing of statins, ezetimibe, or mipomersen for AD prevention. Notwithstanding caveats to this genetic evidence, pharmacovigilance for AD risk among users of PCSK9 inhibitors may be warranted. ANN NEUROL 2020;87:30-39.
© 2019 The Authors. Annals of Neurology published by Wiley Periodicals, Inc. on behalf of American Neurological Association.
Conflict of interest statement
Nothing to report.
Figures


References
-
- Corbett A, Pickett J, Burns A, et al. Drug repositioning for Alzheimer's disease. Nat Rev Drug Discov 2012;11:833–846. - PubMed
-
- Meng X‐F, Yu J‐T, Wang H‐F, et al. Midlife vascular risk factors and the risk of Alzheimer's disease: a systematic review and meta‐analysis. J Alzheimers Dis 2014;42:1295–1310. - PubMed
-
- Matsuzaki T, Sasaki K, Hata J, et al. Association of Alzheimer disease pathology with abnormal lipid metabolism: the Hisayama study. Neurology 2011;77:1068–1075. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

