A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo
- PMID: 31715132
- PMCID: PMC6880868
- DOI: 10.1016/j.ccell.2019.10.002
A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo
Abstract
Signal transducer and activator of transcription 3 (STAT3) is an attractive cancer therapeutic target. Here we report the discovery of SD-36, a small-molecule degrader of STAT3. SD-36 potently induces the degradation of STAT3 protein in vitro and in vivo and demonstrates high selectivity over other STAT members. Induced degradation of STAT3 results in a strong suppression of its transcription network in leukemia and lymphoma cells. SD-36 inhibits the growth of a subset of acute myeloid leukemia and anaplastic large-cell lymphoma cell lines by inducing cell-cycle arrest and/or apoptosis. SD-36 achieves complete and long-lasting tumor regression in multiple xenograft mouse models at well-tolerated dose schedules. Degradation of STAT3 protein, therefore, is a promising cancer therapeutic strategy.
Keywords: PROTAC; SH2 domain; STAT3; c-Myc; degrader; leukemia; lymphoma; selectivity; transcriptional factor; undruggable.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures



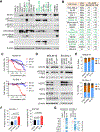
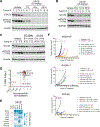

Comment in
-
Inhibit versus Destroy: Are PROTAC Degraders the Solution to Targeting STAT3?Cancer Cell. 2019 Nov 11;36(5):459-461. doi: 10.1016/j.ccell.2019.10.010. Cancer Cell. 2019. PMID: 31715126
-
Dumping STAT3 in the trash.Nat Rev Drug Discov. 2020 Jan;19(1):19. doi: 10.1038/d41573-019-00197-3. Nat Rev Drug Discov. 2020. PMID: 31907421 No abstract available.
References
-
- Becker S, Groner B, and Muller CW (1998). Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 394, 145–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

