Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India
- PMID: 31719139
- PMCID: PMC7407896
- DOI: 10.1128/mSystems.00438-19
Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India
Abstract
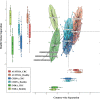
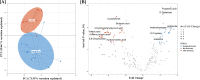
Recently, dysbiosis in the human gut microbiome and shifts in the relative abundances of several bacterial species have been recognized as important factors in colorectal cancer (CRC). However, these studies have been carried out mainly in developed countries where CRC has a high incidence, and it is unclear whether the host-microbiome relationships deduced from these studies can be generalized to the global population. To test if the documented associations between the microbiome and CRC are conserved in a distinct context, we performed metagenomic and metabolomic association studies on fecal samples from 30 CRC patients and 30 healthy controls from two different locations in India, followed by a comparison of CRC data available from other populations. We confirmed the association of Bacteroides and other bacterial taxa with CRC that have been previously reported in other studies. However, the association of CRC with Flavonifractor plautii in Indian patients emerged as a novel finding. The plausible role of F. plautii appears to be linked with the degradation of beneficial anticarcinogenic flavonoids, which was also found to be significantly correlated with the enzymes and modules involved in flavonoid degradation within Indian CRC samples. Thus, we hypothesize that the degradation of beneficial flavonoids might be playing a role in cancer progression within this Indian cohort. We also identified 20 potential microbial taxonomic markers and 33 potential microbial gene markers that discriminate the Indian CRC from healthy microbiomes with high accuracy based on machine learning approaches.IMPORTANCE This study provides novel insights on the CRC-associated microbiome of a unique cohort in India, reveals the potential role of a new bacterium in CRC, and identifies cohort-specific biomarkers, which can potentially be used in noninvasive diagnosis of CRC. The study gains additional significance, as India is among the countries with a very low incidence of CRC, and the diet and lifestyle in India have been associated with a distinct gut microbiome in healthy Indians compared to other global populations. Thus, in this study, we hypothesize a unique relationship between CRC and the gut microbiome in an Indian population.
Keywords: Flavonifractor plautii; biomarkers; colorectal cancer; gut microbiome.
Copyright © 2019 Gupta et al.
Figures




References
-
- Saxena R, Sharma V. 2015. A metagenomic insight into the human microbiome: its implications in health and disease, p 107–119. In Kumar D, Antonarakis S (ed), Medical and health genomics. Elsevier, New York, NY.
-
- Maji A, Misra R, Dhakan DB, Gupta V, Mahato NK, Saxena R, Mittal P, Thukral N, Sharma E, Singh A, Virmani R, Gaur M, Singh H, Hasija Y, Arora G, Agrawal A, Chaudhry A, Khurana JP, Sharma VK, Lal R, Singh Y. 2018. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol 20:402–419. doi:10.1111/1462-2920.14015. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
