High-Risk International Clones of Carbapenem-Nonsusceptible Pseudomonas aeruginosa Endemic to Indonesian Intensive Care Units: Impact of a Multifaceted Infection Control Intervention Analyzed at the Genomic Level
- PMID: 31719179
- PMCID: PMC6851282
- DOI: 10.1128/mBio.02384-19
High-Risk International Clones of Carbapenem-Nonsusceptible Pseudomonas aeruginosa Endemic to Indonesian Intensive Care Units: Impact of a Multifaceted Infection Control Intervention Analyzed at the Genomic Level
Erratum in
-
Erratum for Pelegrin et al., "High-Risk International Clones of Carbapenem-Nonsusceptible Pseudomonas aeruginosa Endemic to Indonesian Intensive Care Units: Impact of a Multifaceted Infection Control Intervention Analyzed at the Genomic Level".mBio. 2020 Jun 16;11(3):e01372-20. doi: 10.1128/mBio.01372-20. mBio. 2020. PMID: 32546610 Free PMC article. No abstract available.
Abstract
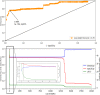
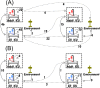
Infection control effectiveness evaluations require detailed epidemiological and microbiological data. We analyzed the genomic profiles of carbapenem-nonsusceptible Pseudomonas aeruginosa (CNPA) strains collected from two intensive care units (ICUs) in the national referral hospital in Jakarta, Indonesia, where a multifaceted infection control intervention was applied. We used clinical data combined with whole-genome sequencing (WGS) of systematically collected CNPA to infer the transmission dynamics of CNPA strains and to characterize their resistome. We found that the number of CNPA transmissions and acquisitions by patients was highly variable over time but that, overall, the rates were not significantly reduced by the intervention. Environmental sources were involved in these transmissions and acquisitions. Four high-risk international CNPA clones (ST235, ST823, ST357, and ST446) dominated, but the distribution of these clones changed significantly after the intervention was implemented. Using resistome analysis, carbapenem resistance was explained by the presence of various carbapenemase-encoding genes (blaGES-5, blaVIM-2-8, and blaIMP-1-7-43) and by mutations within the porin OprD. Our results reveal for the first time the dynamics of P. aeruginosa antimicrobial resistance (AMR) profiles in Indonesia and additionally show the utility of WGS in combination with clinical data to evaluate the impact of an infection control intervention. (This study has been registered at www.trialregister.nl under registration no. NTR5541).IMPORTANCE In low-to-middle-income countries such as Indonesia, work in intensive care units (ICUs) can be hampered by lack of resources. Conducting large epidemiological studies in such settings using genomic tools is rather challenging. Still, we were able to systematically study the transmissions of carbapenem-nonsusceptible strains of P. aeruginosa (CNPA) within and between ICUs, before and after an infection control intervention. Our data show the importance of the broad dissemination of the internationally recognized CNPA clones, the relevance of environmental reservoirs, and the mixed effects of the implemented intervention; it led to a profound change in the clonal make-up of CNPA, but it did not reduce the patients' risk of CNPA acquisitions. Thus, CNPA epidemiology in Indonesian ICUs is part of a global expansion of multiple CNPA clones that remains difficult to control by infection prevention measures.
Keywords: Indonesia; Pseudomonas aeruginosa; infection control; intensive care units; microbial drug resistance; single nucleotide polymorphism.
Copyright © 2019 Pelegrin et al.
Figures


References
-
- European Centre for Disease Prevention and Control. 2018. Surveillance of antimicrobial resistance in Europe 2017. Annual report of the European Antimicrobial Resistance Network (EARS-Net). https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobi....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
