Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians
- PMID: 31719197
- PMCID: PMC6883788
- DOI: 10.1073/pnas.1907883116
Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians
Abstract
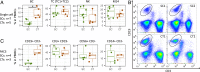
Supercentenarians, people who have reached 110 y of age, are a great model of healthy aging. Their characteristics of delayed onset of age-related diseases and compression of morbidity imply that their immune system remains functional. Here we performed single-cell transcriptome analysis of 61,202 peripheral blood mononuclear cells (PBMCs), derived from 7 supercentenarians and 5 younger controls. We identified a marked increase of cytotoxic CD4 T cells (CD4 cytotoxic T lymphocytes [CTLs]) as a signature of supercentenarians. Furthermore, single-cell T cell receptor sequencing of 2 supercentenarians revealed that CD4 CTLs had accumulated through massive clonal expansion, with the most frequent clonotypes accounting for 15 to 35% of the entire CD4 T cell population. The CD4 CTLs exhibited substantial heterogeneity in their degree of cytotoxicity as well as a nearly identical transcriptome to that of CD8 CTLs. This indicates that CD4 CTLs utilize the transcriptional program of the CD8 lineage while retaining CD4 expression. Indeed, CD4 CTLs extracted from supercentenarians produced IFN-γ and TNF-α upon ex vivo stimulation. Our study reveals that supercentenarians have unique characteristics in their circulating lymphocytes, which may represent an essential adaptation to achieve exceptional longevity by sustaining immune responses to infections and diseases.
Keywords: CD4 CTL; aging; centenarian; single-cell transcriptome.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Young R. D., Validated living worldwide supercentenarians, living and recently deceased: February 2018. Rejuvenation Res. 21, 67–69 (2018). - PubMed
-
- Pavlidis N., Stanta G., Audisio R. A., Cancer prevalence and mortality in centenarians: A systematic review. Crit. Rev. Oncol. Hematol. 83, 145–152 (2012). - PubMed
-
- Willcox D. C., et al. , Life at the extreme limit: Phenotypic characteristics of supercentenarians in Okinawa. J. Gerontol. A Biol. Sci. Med. Sci. 63, 1201–1208 (2008). - PubMed
-
- Evert J., Lawler E., Bogan H., Perls T., Morbidity profiles of centenarians: Survivors, delayers, and escapers. J. Gerontol. A Biol. Sci. Med. Sci. 58, 232–237 (2003). - PubMed
-
- Arai Y., et al. , Physical independence and mortality at the extreme limit of life span: Supercentenarians study in Japan. J. Gerontol. A Biol. Sci. Med. Sci. 69, 486–494 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

