Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis
- PMID: 31719203
- PMCID: PMC6883790
- DOI: 10.1073/pnas.1908364116
Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis
Abstract
Enzymes that generate ribosomally synthesized and posttranslationally modified peptide (RiPP) natural products have garnered significant interest, given their ability to produce large libraries of chemically diverse scaffolds. Such RiPP biosynthetic enzymes are predicted to bind their corresponding peptide substrates through sequence-specific recognition of the leader sequence, which is removed after the installation of posttranslational modifications on the core sequence. The conservation of the leader sequence within a given RiPP class, in otherwise disparate precursor peptides, further supports the notion that strict sequence specificity is necessary for leader peptide engagement. Here, we demonstrate that leader binding by a biosynthetic enzyme in the lasso peptide class of RiPPs is directed by a minimal number of hydrophobic interactions. Biochemical and structural data illustrate how a single leader-binding domain can engage sequence-divergent leader peptides using a conserved motif that facilitates hydrophobic packing. The presence of this simple motif in noncognate peptides results in low micromolar affinity binding by binding domains from several different lasso biosynthetic systems. We also demonstrate that these observations likely extend to other RiPP biosynthetic classes. The portability of the binding motif opens avenues for the engineering of semisynthetic hybrid RiPP products.
Keywords: RiPPs; biochemistry; biosynthesis.
Conflict of interest statement
The authors declare no competing interest.
Figures
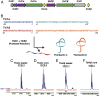
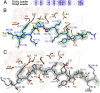

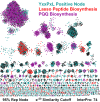

References
-
- Zimmermann M., Hegemann J. D., Xie X., Marahiel M. A., Characterization of caulonodin lasso peptides revealed unprecedented N-terminal residues and a precursor motif essential for peptide maturation. Chem. Sci. (Camb.) 5, 4032–4043 (2014).
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

