Genome-wide association study of eosinophilic granulomatosis with polyangiitis reveals genomic loci stratified by ANCA status
- PMID: 31719529
- PMCID: PMC6851141
- DOI: 10.1038/s41467-019-12515-9
Genome-wide association study of eosinophilic granulomatosis with polyangiitis reveals genomic loci stratified by ANCA status
Abstract
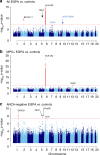
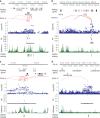
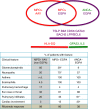
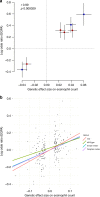
Eosinophilic granulomatosis with polyangiitis (EGPA) is a rare inflammatory disease of unknown cause. 30% of patients have anti-neutrophil cytoplasmic antibodies (ANCA) specific for myeloperoxidase (MPO). Here, we describe a genome-wide association study in 676 EGPA cases and 6809 controls, that identifies 4 EGPA-associated loci through conventional case-control analysis, and 4 additional associations through a conditional false discovery rate approach. Many variants are also associated with asthma and six are associated with eosinophil count in the general population. Through Mendelian randomisation, we show that a primary tendency to eosinophilia contributes to EGPA susceptibility. Stratification by ANCA reveals that EGPA comprises two genetically and clinically distinct syndromes. MPO+ ANCA EGPA is an eosinophilic autoimmune disease sharing certain clinical features and an HLA-DQ association with MPO+ ANCA-associated vasculitis, while ANCA-negative EGPA may instead have a mucosal/barrier dysfunction origin. Four candidate genes are targets of therapies in development, supporting their exploration in EGPA.
Conflict of interest statement
S.L. is a partner in Peptide Groove LLP. The remaining authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

