The gene structure and hypervariability of the complete Penaeus monodon Dscam gene
- PMID: 31719551
- PMCID: PMC6851185
- DOI: 10.1038/s41598-019-52656-x
The gene structure and hypervariability of the complete Penaeus monodon Dscam gene
Abstract
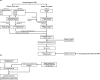
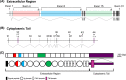
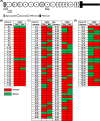
Using two advanced sequencing approaches, Illumina and PacBio, we derive the entire Dscam gene from an M2 assembly of the complete Penaeus monodon genome. The P. monodon Dscam (PmDscam) gene is ~266 kbp, with a total of 44 exons, 5 of which are subject to alternative splicing. PmDscam has a conserved architectural structure consisting of an extracellular region with hypervariable Ig domains, a transmembrane domain, and a cytoplasmic tail. We show that, contrary to a previous report, there are in fact 26, 81 and 26 alternative exons in N-terminal Ig2, N-terminal Ig3 and the entirety of Ig7, respectively. We also identified two alternatively spliced exons in the cytoplasmic tail, with transmembrane domains in exon variants 32.1 and 32.2, and stop codons in exon variants 44.1 and 44.2. This means that alternative splicing is involved in the selection of the stop codon. There are also 7 non-constitutive cytoplasmic tail exons that can either be included or skipped. Alternative splicing and the non-constitutive exons together produce more than 21 million isoform combinations from one PmDscam locus in the P. monodon gene. A public-facing database that allows BLAST searches of all 175 exons in the PmDscam gene has been established at http://pmdscam.dbbs.ncku.edu.tw/ .
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

