Biodiversity of protists and nematodes in the wild nonhuman primate gut
- PMID: 31719654
- PMCID: PMC6976604
- DOI: 10.1038/s41396-019-0551-4
Biodiversity of protists and nematodes in the wild nonhuman primate gut
Abstract
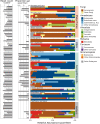
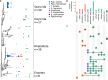
Documenting the natural diversity of eukaryotic organisms in the nonhuman primate (NHP) gut is important for understanding the evolution of the mammalian gut microbiome, its role in digestion, health and disease, and the consequences of anthropogenic change on primate biology and conservation. Despite the ecological significance of gut-associated eukaryotes, little is known about the factors that influence their assembly and diversity in mammals. In this study, we used an 18S rRNA gene fragment metabarcoding approach to assess the eukaryotic assemblage of 62 individuals representing 16 NHP species. We find that cercopithecoids, and especially the cercopithecines, have substantially higher alpha diversity than other NHP groups. Gut-associated protists and nematodes are widespread among NHPs, consistent with their ancient association with NHP hosts. However, we do not find a consistent signal of phylosymbiosis or host-species specificity. Rather, gut eukaryotes are only weakly structured by primate phylogeny with minimal signal from diet, in contrast to previous reports of NHP gut bacteria. The results of this study indicate that gut-associated eukaryotes offer different information than gut-associated bacteria and add to our understanding of the structure of the gut microbiome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Hale VL, Tan CL, Niu K, Yang Y, Knight R, Zhang Q, et al. Diet versus phylogeny: a comparison of gut microbiota in captive colobine monkey species. Microb Ecol. 2018;75:515–27. - PubMed
-
- Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol. 2014;23:1301–17. - PubMed

