Optogenetic Repressors of Gene Expression in Yeasts Using Light-Controlled Nuclear Localization
- PMID: 31719930
- PMCID: PMC6816687
- DOI: 10.1007/s12195-019-00598-9
Optogenetic Repressors of Gene Expression in Yeasts Using Light-Controlled Nuclear Localization
Abstract
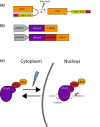
Introduction: Controlling gene expression is a fundamental goal of basic and synthetic biology because it allows insight into cellular function and control of cellular activity. We explored the possibility of generating an optogenetic repressor of gene expression in the model organism Saccharomyces cerevisiae by using light to control the nuclear localization of nuclease-dead Cas9, dCas9.
Methods: The dCas9 protein acts as a repressor for a gene of interest when localized to the nucleus in the presence of an appropriate guide RNA (sgRNA). We engineered dCas9, the mammalian transcriptional repressor Mxi1, and an optogenetic tool to control nuclear localization (LINuS) as parts in an existing yeast optogenetic toolkit. This allowed expression cassettes containing novel dCas9 repressor configurations and guide RNAs to be rapidly constructed and integrated into yeast.
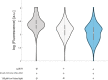
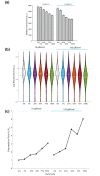
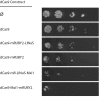
Results: Our library of repressors displays a range of basal repression without the need for inducers or promoter modification. Populations of cells containing these repressors can be combined to generate a heterogeneous population of yeast with a 100-fold expression range. We find that repression can be dialed modestly in a light dose- and intensity-dependent manner. We used this library to repress expression of the lanosterol 14-alpha-demethylase Erg11, generating yeast with a range of sensitivity to the important antifungal drug fluconazole.
Conclusions: This toolkit will be useful for spatiotemporal perturbation of gene expression in Saccharomyces cerevisiae. Additionally, we believe that the simplicity of our scheme will allow these repressors to be easily modified to control gene expression in medically relevant fungi, such as pathogenic yeasts.
Keywords: Fungal drug resistance; Gene expression; LINuS; Optogenetics; dCas9.
© Biomedical Engineering Society 2019.
Figures







References
-
- Aubrey B, Kelly G, Brennan KAM, O’Connor L, Milla L, Wilcox S, Tai L, Strasser A, Herold M. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Reports. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. - DOI - PubMed
-
- Bódi Z, Farkas Z, Nevozhay D, Kalapis D, Lázár V, Csörgö B, Nyerges Á, Szamecz B, Fekete G, Papp B, Araújo H, Oliveira J, Moura G, Santos M, Székely T, Jr, Balázsi G, Pál C. Phenotypic heterogeneity promotes adaptive evolution. PLOS Biology. 2017;15(5):e2000644. doi: 10.1371/journal.pbio.2000644. - DOI - PMC - PubMed
-
- Brachmann C, Davies A, Cost G, Caputo E, Li J, Heiter P, Boeke J. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. - DOI - PubMed
-
- Branch M, Coleman T, Li Y. A Subspace, Interior, and Conjugate Gradient Method for Large-Scale Bound-Constrained Minimization Problems. SIAM Journal of Scientific Computing. 1999;21(1):1–23. doi: 10.1137/S1064827595289108. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

