Divergence dating using mixed effects clock modelling: An application to HIV-1
- PMID: 31720009
- PMCID: PMC6830409
- DOI: 10.1093/ve/vez036
Divergence dating using mixed effects clock modelling: An application to HIV-1
Abstract
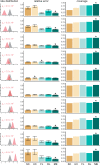
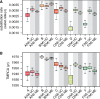
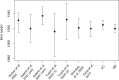
The need to estimate divergence times in evolutionary histories in the presence of various sources of substitution rate variation has stimulated a rich development of relaxed molecular clock models. Viral evolutionary studies frequently adopt an uncorrelated clock model as a generic relaxed molecular clock process, but this may impose considerable estimation bias if discrete rate variation exists among clades or lineages. For HIV-1 group M, rate variation among subtypes has been shown to result in inconsistencies in time to the most recent common ancestor estimation. Although this calls into question the adequacy of available molecular dating methods, no solution to this problem has been offered so far. Here, we investigate the use of mixed effects molecular clock models, which combine both fixed and random effects in the evolutionary rate, to estimate divergence times. Using simulation, we demonstrate that this model outperforms existing molecular clock models in a Bayesian framework for estimating time-measured phylogenies in the presence of mixed sources of rate variation, while also maintaining good performance in simpler scenarios. By analysing a comprehensive HIV-1 group M complete genome data set we confirm considerable rate variation among subtypes that is not adequately modelled by uncorrelated relaxed clock models. The mixed effects clock model can accommodate this rate variation and results in a time to the most recent common ancestor of HIV-1 group M of 1920 (1915-25), which is only slightly earlier than the uncorrelated relaxed clock estimate for the same data set. The use of complete genome data appears to have a more profound impact than the molecular clock model because it reduces the credible intervals by 50 per cent relative to similar estimates based on short envelope gene sequences.
Keywords: Bayesian inference; HIV; divergence time; mixed effects; molecular clock.
© The Author(s) 2019. Published by Oxford University Press.
Figures




Similar articles
-
A mixed relaxed clock model.Philos Trans R Soc Lond B Biol Sci. 2016 Jul 19;371(1699):20150132. doi: 10.1098/rstb.2015.0132. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27325829 Free PMC article.
-
Characterization of the uncertainty of divergence time estimation under relaxed molecular clock models using multiple loci.Syst Biol. 2015 Mar;64(2):267-80. doi: 10.1093/sysbio/syu109. Epub 2014 Dec 11. Syst Biol. 2015. PMID: 25503979 Free PMC article.
-
Rate variation and estimation of divergence times using strict and relaxed clocks.BMC Evol Biol. 2011 Sep 26;11:271. doi: 10.1186/1471-2148-11-271. BMC Evol Biol. 2011. PMID: 21943087 Free PMC article.
-
Modeling Substitution Rate Evolution across Lineages and Relaxing the Molecular Clock.Genome Biol Evol. 2024 Sep 3;16(9):evae199. doi: 10.1093/gbe/evae199. Genome Biol Evol. 2024. PMID: 39332907 Free PMC article. Review.
-
Molecular-clock methods for estimating evolutionary rates and timescales.Mol Ecol. 2014 Dec;23(24):5947-65. doi: 10.1111/mec.12953. Epub 2014 Oct 30. Mol Ecol. 2014. PMID: 25290107 Review.
Cited by
-
Phylogenetic analysis of HIV-1 shows frequent cross-country transmission and local population expansions.Virus Evol. 2021 Jun 9;7(2):veab055. doi: 10.1093/ve/veab055. eCollection 2021. Virus Evol. 2021. PMID: 34532059 Free PMC article.
-
Inferring Viral Transmission Time from Phylogenies for Known Transmission Pairs.Mol Biol Evol. 2024 Jan 3;41(1):msad282. doi: 10.1093/molbev/msad282. Mol Biol Evol. 2024. PMID: 38149995 Free PMC article.
-
Newly identified lineages of porcine hemagglutinating encephalomyelitis virus exhibit respiratory phenotype.Virus Evol. 2023 Aug 18;9(2):vead051. doi: 10.1093/ve/vead051. eCollection 2023. Virus Evol. 2023. PMID: 37711483 Free PMC article.
-
Mapping Transmission Dynamics and Drug Resistance Surveillance in the Cyprus HIV-1 Epidemic (2017-2021).Viruses. 2024 Sep 11;16(9):1449. doi: 10.3390/v16091449. Viruses. 2024. PMID: 39339925 Free PMC article.
-
Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil.medRxiv [Preprint]. 2021 Mar 3:2021.02.26.21252554. doi: 10.1101/2021.02.26.21252554. medRxiv. 2021. Update in: Science. 2021 May 21;372(6544):815-821. doi: 10.1126/science.abh2644. PMID: 33688664 Free PMC article. Updated. Preprint.

