Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis
- PMID: 31720123
- PMCID: PMC6842560
- DOI: 10.7717/peerj.8019
Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis
Abstract
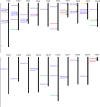
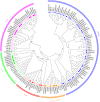
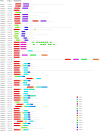
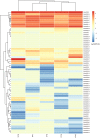
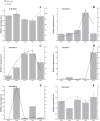
MADS-box genes encode transcription factors that participate in various plant growth and development processes, particularly floral organogenesis. To date, MADS-box genes have been reported in many species, the completion of the sequence of the willow genome provides us with the opportunity to conduct a comprehensive analysis of the willow MADS-box gene family. Here, we identified 60 willow MADS-box genes using bioinformatics-based methods and classified them into 22 M-type (11 Mα, seven Mβ and four Mγ) and 38 MIKC-type (32 MIKCc and six MIKC*) genes based on a phylogenetic analysis. Fifty-six of the 60 SsMADS genes were randomly distributed on 19 putative willow chromosomes. By combining gene structure analysis with evolutionary analysis, we found that the MIKC-type genes were more conserved and played a more important role in willow growth. Further study showed that the MIKC* type was a transition between the M-type and MIKC-type. Additionally, the number of MADS-box genes in gymnosperms was notably lower than that in angiosperms. Finally, the expression profiles of these willow MADS-box genes were analysed in five different tissues (root, stem, leave, bud and bark) and validated by RT-qPCR experiments. This study is the first genome-wide analysis of the willow MADS-box gene family, and the results establish a basis for further functional studies of willow MADS-box genes and serve as a reference for related studies of other woody plants.
Keywords: Expression; Gene family; Genome-wide characterization; MADS-box; Phylogenetic analysis; Willow.
©2019 Qu et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A.BMC Genomics. 2018 Oct 3;19(1):726. doi: 10.1186/s12864-018-5113-z. BMC Genomics. 2018. PMID: 30285611 Free PMC article.
-
Structural and functional annotation of the MADS-box transcription factor family in grapevine.BMC Genomics. 2016 Jan 27;17:80. doi: 10.1186/s12864-016-2398-7. BMC Genomics. 2016. PMID: 26818751 Free PMC article.
-
MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress.BMC Genomics. 2007 Jul 18;8:242. doi: 10.1186/1471-2164-8-242. BMC Genomics. 2007. PMID: 17640358 Free PMC article.
-
Genome-wide Analysis of the MADS-Box Gene Family in Watermelon.Comput Biol Chem. 2019 Jun;80:341-350. doi: 10.1016/j.compbiolchem.2019.04.013. Epub 2019 May 2. Comput Biol Chem. 2019. PMID: 31082717 Review.
-
The major clades of MADS-box genes and their role in the development and evolution of flowering plants.Mol Phylogenet Evol. 2003 Dec;29(3):464-89. doi: 10.1016/s1055-7903(03)00207-0. Mol Phylogenet Evol. 2003. PMID: 14615187 Review.
Cited by
-
Comprehensive bioinformation analysis of homeodomain-leucine zipper gene family and expression pattern of HD-Zip I under abiotic stress in Salix suchowensis.BMC Genomics. 2024 Feb 15;25(1):182. doi: 10.1186/s12864-024-10067-x. BMC Genomics. 2024. PMID: 38360569 Free PMC article.
-
Identification and Expression Analysis of the Populus trichocarpa GASA-Gene Family.Int J Mol Sci. 2022 Jan 28;23(3):1507. doi: 10.3390/ijms23031507. Int J Mol Sci. 2022. PMID: 35163431 Free PMC article.
-
Genome-Wide Identification of the Ginkgo (Ginkgo biloba L.) LBD Transcription Factor Gene and Characterization of Its Expression.Int J Mol Sci. 2022 May 13;23(10):5474. doi: 10.3390/ijms23105474. Int J Mol Sci. 2022. PMID: 35628284 Free PMC article.
-
Genome-Wide Identification of PLATZ Transcription Factors in Ginkgo biloba L. and Their Expression Characteristics During Seed Development.Front Plant Sci. 2022 Jun 23;13:946194. doi: 10.3389/fpls.2022.946194. eCollection 2022. Front Plant Sci. 2022. PMID: 35812908 Free PMC article.
-
Identification and Expression of the MADS-box Gene Family in Different Versions of the Ginkgo biloba Genome.Plants (Basel). 2023 Sep 21;12(18):3334. doi: 10.3390/plants12183334. Plants (Basel). 2023. PMID: 37765498 Free PMC article.
References
-
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas DPL, Martinez-Castilla L, Yanofsky MF. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources

