A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data
- PMID: 31720216
- PMCID: PMC6838544
- DOI: 10.1016/j.mec.2019.e00101
A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data
Erratum in
-
Erratum regarding previously published articles in volumes 9, 10 and 11.Metab Eng Commun. 2021 Oct 28;13:e00186. doi: 10.1016/j.mec.2021.e00186. eCollection 2021 Dec. Metab Eng Commun. 2021. PMID: 34765440 Free PMC article.
Abstract
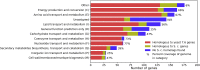
Rhodosporidium toruloides is a red, basidiomycetes yeast that can accumulate a large amount of lipids and produce carotenoids. To better assess this non-model yeast's metabolic capabilities, we reconstructed a genome-scale model of R. toruloides IFO0880's metabolic network (iRhto1108) accounting for 2204 reactions, 1985 metabolites and 1108 genes. In this work, we integrated and supplemented the current knowledge with in-house generated biomass composition and experimental measurements pertaining to the organism's metabolic capabilities. Predictions of genotype-phenotype relations were improved through manual curation of gene-protein-reaction rules for 543 reactions leading to correct recapitulations of 84.5% of gene essentiality data (sensitivity of 94.3% and specificity of 53.8%). Organism-specific macromolecular composition and ATP maintenance requirements were experimentally measured for two separate growth conditions: (i) carbon and (ii) nitrogen limitations. Overall, iRhto1108 reproduced R. toruloides's utilization capabilities for 18 alternate substrates, matched measured wild-type growth yield, and recapitulated the viability of 772 out of 819 deletion mutants. As a demonstration to the model's fidelity in guiding engineering interventions, the OptForce procedure was applied on iRhto1108 for triacylglycerol overproduction. Suggested interventions recapitulated many of the previous successful implementations of genetic modifications and put forth a few new ones.
© 2019 The Authors.
Figures

References
-
- Andrews S. 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc [WWW Document]. URL.
-
- Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S., Sneddon M.W., Henderson M.L., Riehl W.J., Murphy-Olson D., Chan S.Y., Kamimura R.T., Kumari S., Drake M.M., Brettin T.S., Glass E.M., Chivian D., Gunter D., Weston D.J., Allen B.H., Baumohl J., Best A.A., Bowen B., Brenner S.E., Bun C.C., Chandonia J.-M., Chia J.-M., Colasanti R., Conrad N., Davis J.J., Davison B.H., DeJongh M., Devoid S., Dietrich E., Dubchak I., Edirisinghe J.N., Fang G., Faria J.P., Frybarger P.M., Gerlach W., Gerstein M., Greiner A., Gurtowski J., Haun H.L., He F., Jain R., Joachimiak M.P., Keegan K.P., Kondo S., Kumar V., Land M.L., Meyer F., Mills M., Novichkov P.S., Oh T., Olsen G.J., Olson R., Parrello B., Pasternak S., Pearson E., Poon S.S., Price G.A., Ramakrishnan S., Ranjan P., Ronald P.C., Schatz M.C., Seaver S.M.D., Shukla M., Sutormin R.A., Syed M.H., Thomason J., Tintle N.L., Wang D., Xia F., Yoo H., Yoo S., Yu D. KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

