Structure and function relationships in mammalian DNA polymerases
- PMID: 31722068
- PMCID: PMC7050493
- DOI: 10.1007/s00018-019-03368-y
Structure and function relationships in mammalian DNA polymerases
Abstract
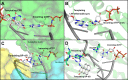
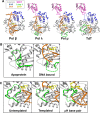
DNA polymerases are vital for the synthesis of new DNA strands. Since the discovery of DNA polymerase I in Escherichia coli, a diverse library of mammalian DNA polymerases involved in DNA replication, DNA repair, antibody generation, and cell checkpoint signaling has emerged. While the unique functions of these DNA polymerases are differentiated by their association with accessory factors and/or the presence of distinctive catalytic domains, atomic resolution structures of DNA polymerases in complex with their DNA substrates have revealed mechanistic subtleties that contribute to their specialization. In this review, the structure and function of all 15 mammalian DNA polymerases from families B, Y, X, and A will be reviewed and discussed with special emphasis on the insights gleaned from recently published atomic resolution structures.
Keywords: DNA polymerase; DNA repair; DNA synthesis; Replication; Structural biology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

