Misregulation of Drosophila Myc Disrupts Circadian Behavior and Metabolism
- PMID: 31722196
- PMCID: PMC6910219
- DOI: 10.1016/j.celrep.2019.10.022
Misregulation of Drosophila Myc Disrupts Circadian Behavior and Metabolism
Abstract
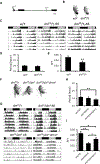
Drosophila Myc (dMyc) is highly conserved and functions as a transcription factor similar to mammalian Myc. We previously found that oncogenic Myc disrupts the molecular clock in cancer cells. Here, we demonstrate that misregulation of dMyc expression affects Drosophila circadian behavior. dMyc overexpression results in a high percentage of arrhythmic flies, concomitant with increases in the expression of clock genes cyc, tim, cry, and cwo. Conversely, flies with hypomorphic mutations in dMyc exhibit considerable arrhythmia, which can be rescued by loss of dMnt, a suppressor of dMyc activity. Metabolic profiling of fly heads revealed that loss of dMyc and its overexpression alter steady-state metabolite levels and have opposing effects on histidine, the histamine precursor, which is rescued in dMyc mutants by ablation of dMnt and could contribute to effects of dMyc on locomotor behavior. Our results demonstrate a role of dMyc in modulating Drosophila circadian clock, behavior, and metabolism.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


References
-
- Abe H, Honma S, Ohtsu H, and Honma K (2004). Circadian rhythms in behavior and clock gene expressions in the brain of mice lacking histidine decarboxylase. Brain Res. Mol. Brain Res 124, 178–187. - PubMed
-
- Asher G, and Schibler U (2011). Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 13, 125–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

