Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis
- PMID: 31722201
- PMCID: PMC6856722
- DOI: 10.1016/j.celrep.2019.10.024
Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis
Abstract
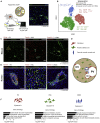
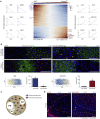
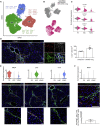
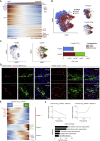
Iterative liver injury results in progressive fibrosis disrupting hepatic architecture, regeneration potential, and liver function. Hepatic stellate cells (HSCs) are a major source of pathological matrix during fibrosis and are thought to be a functionally homogeneous population. Here, we use single-cell RNA sequencing to deconvolve the hepatic mesenchyme in healthy and fibrotic mouse liver, revealing spatial zonation of HSCs across the hepatic lobule. Furthermore, we show that HSCs partition into topographically diametric lobule regions, designated portal vein-associated HSCs (PaHSCs) and central vein-associated HSCs (CaHSCs). Importantly we uncover functional zonation, identifying CaHSCs as the dominant pathogenic collagen-producing cells in a mouse model of centrilobular fibrosis. Finally, we identify LPAR1 as a therapeutic target on collagen-producing CaHSCs, demonstrating that blockade of LPAR1 inhibits liver fibrosis in a rodent NASH model. Taken together, our work illustrates the power of single-cell transcriptomics to resolve the key collagen-producing cells driving liver fibrosis with high precision.
Keywords: hepatic stellate cells; liver fibrosis; mesenchyme; single-cell RNA sequencing; zonation.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
A.Z., S.V.P., T.E.H., M.M.W., G.R.B., and D.G.B. are employees and shareholders of Gilead Sciences, Inc.
Figures




References
-
- Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K.W., Schindelin J., Cardona A., Sebastian Seung H. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B. 1995;57:289–300.
-
- Bhunchet E., Wake K. Role of mesenchymal cell populations in porcine serum-induced rat liver fibrosis. Hepatology. 1992;16:1452–1473. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

