MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data
- PMID: 31722416
- PMCID: PMC7145535
- DOI: 10.1093/nar/gkz1010
MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data
Abstract
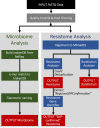
Antimicrobial resistance (AMR) is a threat to global public health and the identification of genetic determinants of AMR is a critical component to epidemiological investigations. High-throughput sequencing (HTS) provides opportunities for investigation of AMR across all microbial genomes in a sample (i.e. the metagenome). Previously, we presented MEGARes, a hand-curated AMR database and annotation structure developed to facilitate the analysis of AMR within metagenomic samples (i.e. the resistome). Along with MEGARes, we released AmrPlusPlus, a bioinformatics pipeline that interfaces with MEGARes to identify and quantify AMR gene accessions contained within a metagenomic sequence dataset. Here, we present MEGARes 2.0 (https://megares.meglab.org), which incorporates previously published resistance sequences for antimicrobial drugs, while also expanding to include published sequences for metal and biocide resistance determinants. In MEGARes 2.0, the nodes of the acyclic hierarchical ontology include four antimicrobial compound types, 57 classes, 220 mechanisms of resistance, and 1,345 gene groups that classify the 7,868 accessions. In addition, we present an updated version of AmrPlusPlus (AMR ++ version 2.0), which improves accuracy of classifications, as well as expanding scalability and usability.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Huttner A., Harbarth S., Carlet J., Cosgrove S., Goossens H., Holmes A., Jarlier V., Voss A., Pittet D. for the World Healthcare-Associated Infections Forum participants Antimicrobial resistance: a global view from the 2013 World Healthcare-Associated Infections Forum. Antimicrob. Resist. Infect. Control. 2013; 2:31. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

