Chromatin accessibility and transcription dynamics during in vitro astrocyte differentiation of Huntington's Disease Monkey pluripotent stem cells
- PMID: 31722751
- PMCID: PMC6852955
- DOI: 10.1186/s13072-019-0313-6
Chromatin accessibility and transcription dynamics during in vitro astrocyte differentiation of Huntington's Disease Monkey pluripotent stem cells
Abstract
Background: Huntington's Disease (HD) is a fatal neurodegenerative disorder caused by a CAG repeat expansion, resulting in a mutant huntingtin protein. While it is now clear that astrocytes are affected by HD and significantly contribute to neuronal dysfunction and pathogenesis, the alterations in the transcriptional and epigenetic profiles in HD astrocytes have yet to be characterized. Here, we examine global transcription and chromatin accessibility dynamics during in vitro astrocyte differentiation in a transgenic non-human primate model of HD.
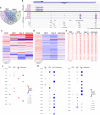
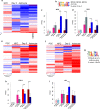
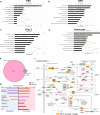
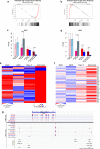
Results: We found global changes in accessibility and transcription across different stages of HD pluripotent stem cell differentiation, with distinct trends first observed in neural progenitor cells (NPCs), once cells have committed to a neural lineage. Transcription of p53 signaling and cell cycle pathway genes was highly impacted during differentiation, with depletion in HD NPCs and upregulation in HD astrocytes. E2F target genes also displayed this inverse expression pattern, and strong associations between E2F target gene expression and accessibility at nearby putative enhancers were observed.
Conclusions: The results suggest that chromatin accessibility and transcription are altered throughout in vitro HD astrocyte differentiation and provide evidence that E2F dysregulation contributes to aberrant cell-cycle re-entry and apoptosis throughout the progression from NPCs to astrocytes.
Keywords: ATAC-seq; Brain; Glia; Neural progenitor cells; Neurodegeneration.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. - DOI - PMC - PubMed
-
- Hu J, Dodd DW, Hudson RH, Corey DR. Cellular localization and allele-selective inhibition of mutant huntingtin protein by peptide nucleic acid oligomers containing the fluorescent nucleobase [bis-o-(aminoethoxy)phenyl]pyrrolocytosine. Bioorg Med Chem Lett. 2009;19:6181–6184. doi: 10.1016/j.bmcl.2009.09.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

