The Global Ascendency of OXA-48-Type Carbapenemases
- PMID: 31722889
- PMCID: PMC6860007
- DOI: 10.1128/CMR.00102-19
The Global Ascendency of OXA-48-Type Carbapenemases
Abstract
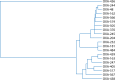
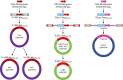
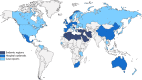
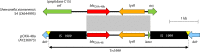
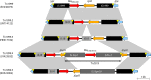
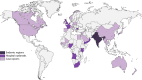
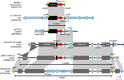
Surveillance studies have shown that OXA-48-like carbapenemases are the most common carbapenemases in Enterobacterales in certain regions of the world and are being introduced on a regular basis into regions of nonendemicity, where they are responsible for nosocomial outbreaks. OXA-48, OXA-181, OXA-232, OXA-204, OXA-162, and OXA-244, in that order, are the most common enzymes identified among the OXA-48-like carbapenemase group. OXA-48 is associated with different Tn1999 variants on IncL plasmids and is endemic in North Africa and the Middle East. OXA-162 and OXA-244 are derivatives of OXA-48 and are present in Europe. OXA-181 and OXA-232 are associated with ISEcp1, Tn2013 on ColE2, and IncX3 types of plasmids and are endemic in the Indian subcontinent (e.g., India, Bangladesh, Pakistan, and Sri Lanka) and certain sub-Saharan African countries. Overall, clonal dissemination plays a minor role in the spread of OXA-48-like carbapenemases, but certain high-risk clones (e.g., Klebsiella pneumoniae sequence type 147 [ST147], ST307, ST15, and ST14 and Escherichia coli ST38 and ST410) have been associated with the global dispersion of OXA-48, OXA-181, OXA-232, and OXA-204. Chromosomal integration of blaOXA-48 within Tn6237 occurred among E. coli ST38 isolates, especially in the United Kingdom. The detection of Enterobacterales with OXA-48-like enzymes using phenotypic methods has improved recently but remains challenging for clinical laboratories in regions of nonendemicity. Identification of the specific type of OXA-48-like enzyme requires sequencing of the corresponding genes. Bacteria (especially K. pneumoniae and E. coli) with blaOXA-48, blaOXA-181, and blaOXA-232 are emerging in different parts of the world and are most likely underreported due to problems with the laboratory detection of these enzymes. The medical community should be aware of the looming threat that is posed by bacteria with OXA-48-like carbapenemases.
Keywords: Enterobacteriaceae; OXA-48-like; carbapenemases.
Copyright © 2019 American Society for Microbiology.
Figures




References
-
- Marchaim D, Gottesman T, Schwartz O, Korem M, Maor Y, Rahav G, Karplus R, Lazarovitch T, Braun E, Sprecher H, Lachish T, Wiener-Well Y, Alon D, Chowers M, Ciobotaro P, Bardenstein R, Paz A, Potasman I, Giladi M, Schechner V, Schwaber MJ, Klarfeld-Lidji S, Carmeli Y. 2010. National multicenter study of predictors and outcomes of bacteremia upon hospital admission caused by Enterobacteriaceae producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother 54:5099–5104. doi:10.1128/AAC.00565-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

