A Metabolic Dependency for Host Isoprenoids in the Obligate Intracellular Pathogen Rickettsia parkeri Underlies a Sensitivity to the Statin Class of Host-Targeted Therapeutics
- PMID: 31722991
- PMCID: PMC6854040
- DOI: 10.1128/mSphere.00536-19
A Metabolic Dependency for Host Isoprenoids in the Obligate Intracellular Pathogen Rickettsia parkeri Underlies a Sensitivity to the Statin Class of Host-Targeted Therapeutics
Erratum in
-
Erratum for Ahyong et al., "A Metabolic Dependency for Host Isoprenoids in the Obligate Intracellular Pathogen Rickettsia parkeri Underlies a Sensitivity to the Statin Class of Host-Targeted Therapeutics".mSphere. 2019 Nov 27;4(6):e00848-19. doi: 10.1128/mSphere.00848-19. mSphere. 2019. PMID: 31776245 Free PMC article. No abstract available.
Abstract
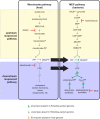
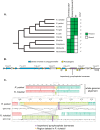
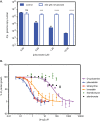
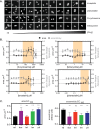
Gram-negative bacteria in the order Rickettsiales have an obligate intracellular growth requirement, and some species cause human diseases such as typhus and spotted fever. The bacteria have evolved a dependence on essential nutrients and metabolites from the host cell as a consequence of extensive genome reduction. However, it remains largely unknown which nutrients they acquire and whether their metabolic dependency can be exploited therapeutically. Here, we describe a genetic rewiring of bacterial isoprenoid biosynthetic pathways in the Rickettsiales that has resulted from reductive genome evolution. Furthermore, we investigated whether the spotted fever group Rickettsia species Rickettsia parkeri scavenges isoprenoid precursors directly from the host. Using targeted mass spectrometry, we found that infection caused decreases in host isoprenoid products and concomitant increases in bacterial isoprenoid metabolites. Additionally, we report that treatment of infected cells with statins, which inhibit host isoprenoid synthesis, prohibited bacterial growth. We show that growth inhibition correlates with changes in bacterial size and shape that mimic those caused by antibiotics that inhibit peptidoglycan biosynthesis, suggesting that statins lead to an inhibition of cell wall synthesis. Altogether, our results describe a potential Achilles' heel of obligate intracellular pathogens that can potentially be exploited with host-targeted therapeutics that interfere with metabolic pathways required for bacterial growth.IMPORTANCE Obligate intracellular pathogens, which include viruses as well as certain bacteria and eukaryotes, are a subset of infectious microbes that are metabolically dependent on and unable to grow outside an infected host cell because they have lost or lack essential biosynthetic pathways. In this study, we describe a metabolic dependency of the bacterial pathogen Rickettsia parkeri on host isoprenoid molecules that are used in the biosynthesis of downstream products, including cholesterol, steroid hormones, and heme. Bacteria make products from isoprenoids, such as an essential lipid carrier for making the bacterial cell wall. We show that bacterial metabolic dependency can represent a potential Achilles' heel and that inhibiting host isoprenoid biosynthesis with the FDA-approved statin class of drugs inhibits bacterial growth by interfering with the integrity of the cell wall. This work supports the potential to treat infections by obligate intracellular pathogens through inhibition of host biosynthetic pathways that are susceptible to parasitism.
Keywords: Rickettsia; isoprenoids; metabolic parasitism.
Copyright © 2019 Ahyong et al.
Figures

References
-
- Yu X-J, Walker DH. 2006. The order Rickettsiales, p 493–528. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The Prokaryotes. Springer, New York, NY.
-
- Paddock CD, Finley RW, Wright CS, Robinson HN, Schrodt BJ, Lane CC, Ekenna O, Blass MA, Tamminga CL, Ohl CA, McLellan SLF, Goddard J, Holman RC, Openshaw JJ, Sumner JW, Zaki SR, Eremeeva ME. 2008. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin Infect Dis 47:1188–1196. doi:10.1086/592254. - DOI - PubMed

