Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors
- PMID: 31723052
- PMCID: PMC6948855
- DOI: 10.1172/jci.insight.131610
Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors
Abstract
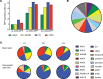
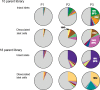
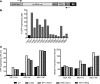
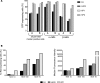
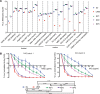
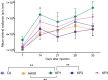
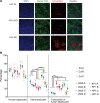
While gene transfer using recombinant adeno-associated viral (rAAV) vectors has shown success in some clinical trials, there remain many tissues that are not well transduced. Because of the recent success in reprogramming islet-derived cells into functional β cells in animal models, we constructed 2 highly complex barcoded replication competent capsid shuffled libraries and selected for high-transducing variants on primary human islets. We describe the generation of a chimeric AAV capsid (AAV-KP1) that facilitates transduction of primary human islet cells and human embryonic stem cell-derived β cells with up to 10-fold higher efficiency compared with previously studied best-in-class AAV vectors. Remarkably, this chimeric capsid also enabled transduction of both mouse and human hepatocytes at very high levels in a humanized chimeric mouse model, thus providing a versatile vector that has the potential to be used in both preclinical testing and human clinical trials for liver-based diseases and diabetes.
Keywords: Diabetes; Embryonic stem cells; Gene therapy; Therapeutics.
Conflict of interest statement
Figures

References
-
- CDC. US Department of Health Human Services. Web Site. https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-stat... Updated 2017. Accessed October 30, 2019.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

