Clonal selection confers distinct evolutionary trajectories in BRAF-driven cancers
- PMID: 31723142
- PMCID: PMC6853924
- DOI: 10.1038/s41467-019-13161-x
Clonal selection confers distinct evolutionary trajectories in BRAF-driven cancers
Abstract
Molecular determinants governing the evolution of tumor subclones toward phylogenetic branches or fixation remain unknown. Using sequencing data, we model the propagation and selection of clones expressing distinct categories of BRAF mutations to estimate their evolutionary trajectories. We show that strongly activating BRAF mutations demonstrate hard sweep dynamics, whereas mutations with less pronounced activation of the BRAF signaling pathway confer soft sweeps or are subclonal. We use clonal reconstructions to estimate the strength of "driver" selection in individual tumors. Using tumors cells and human-derived murine xenografts, we show that tumor sweep dynamics can significantly affect responses to targeted inhibitors of BRAF/MEK or DNA damaging agents. Our study uncovers patterns of distinct BRAF clonal evolutionary dynamics and nominates therapeutic strategies based on the identity of the BRAF mutation and its clonal composition.
Conflict of interest statement
M.E.A. receives grant support, travel support, and honoraria from Bayer AG and receives grant support from Siemens Medical Solutions, USA in subject matter or material not directly related to this work. The other authors disclose no potential conflicts of interest.
Figures

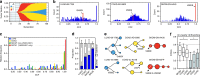
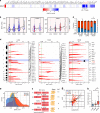
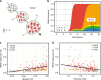
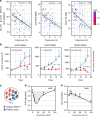

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

