The neXtProt knowledgebase in 2020: data, tools and usability improvements
- PMID: 31724716
- PMCID: PMC7145669
- DOI: 10.1093/nar/gkz995
The neXtProt knowledgebase in 2020: data, tools and usability improvements
Abstract
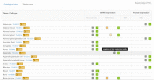
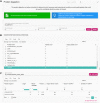
The neXtProt knowledgebase (https://www.nextprot.org) is an integrative resource providing both data on human protein and the tools to explore these. In order to provide comprehensive and up-to-date data, we evaluate and add new data sets. We describe the incorporation of three new data sets that provide expression, function, protein-protein binary interaction, post-translational modifications (PTM) and variant information. New SPARQL query examples illustrating uses of the new data were added. neXtProt has continued to develop tools for proteomics. We have improved the peptide uniqueness checker and have implemented a new protein digestion tool. Together, these tools make it possible to determine which proteases can be used to identify trypsin-resistant proteins by mass spectrometry. In terms of usability, we have finished revamping our web interface and completely rewritten our API. Our SPARQL endpoint now supports federated queries. All the neXtProt data are available via our user interface, API, SPARQL endpoint and FTP site, including the new PEFF 1.0 format files. Finally, the data on our FTP site is now CC BY 4.0 to promote its reuse.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. et al. .. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

