Development of Genome-Derived Tumor Type Prediction to Inform Clinical Cancer Care
- PMID: 31725847
- PMCID: PMC6865333
- DOI: 10.1001/jamaoncol.2019.3985
Development of Genome-Derived Tumor Type Prediction to Inform Clinical Cancer Care
Abstract
Importance: Diagnosing the site of origin for cancer is a pillar of disease classification that has directed clinical care for more than a century. Even in an era of precision oncologic practice, in which treatment is increasingly informed by the presence or absence of mutant genes responsible for cancer growth and progression, tumor origin remains a critical factor in tumor biologic characteristics and therapeutic sensitivity.
Objective: To evaluate whether data derived from routine clinical DNA sequencing of tumors could complement conventional approaches to enable improved diagnostic accuracy.
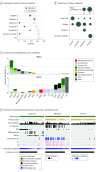
Design, setting, and participants: A machine learning approach was developed to predict tumor type from targeted panel DNA sequence data obtained at the point of care, incorporating both discrete molecular alterations and inferred features such as mutational signatures. This algorithm was trained on 7791 tumors representing 22 cancer types selected from a prospectively sequenced cohort of patients with advanced cancer.
Results: The correct tumor type was predicted for 5748 of the 7791 patients (73.8%) in the training set as well as 8623 of 11 644 patients (74.1%) in an independent cohort. Predictions were assigned probabilities that reflected empirical accuracy, with 3388 cases (43.5%) representing high-confidence predictions (>95% probability). Informative molecular features and feature categories varied widely by tumor type. Genomic analysis of plasma cell-free DNA yielded accurate predictions in 45 of 60 cases (75.0%), suggesting that this approach may be applied in diverse clinical settings including as an adjunct to cancer screening. Likely tissues of origin were predicted from targeted tumor sequencing in 95 of 141 patients (67.4%) with cancers of unknown primary site. Applying this method prospectively to patients under active care enabled genome-directed reassessment of diagnosis in 2 patients initially presumed to have metastatic breast cancer, leading to the selection of more appropriate treatments, which elicited clinical responses.
Conclusions and relevance: These results suggest that the application of artificial intelligence to predict tissue of origin in oncologic practice can act as a useful complement to conventional histologic review to provide integrated pathologic diagnoses, often with important therapeutic implications.
Conflict of interest statement
Figures

Comment in
-
Tumor Origins Through Genomic Profiles.JAMA Oncol. 2020 Jan 1;6(1):33-34. doi: 10.1001/jamaoncol.2019.3981. JAMA Oncol. 2020. PMID: 31725824 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

