Investigation of Molecular Details of Keap1-Nrf2 Inhibitors Using Molecular Dynamics and Umbrella Sampling Techniques
- PMID: 31726716
- PMCID: PMC6891428
- DOI: 10.3390/molecules24224085
Investigation of Molecular Details of Keap1-Nrf2 Inhibitors Using Molecular Dynamics and Umbrella Sampling Techniques
Abstract
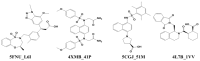
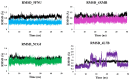
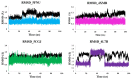
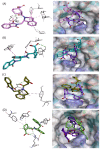
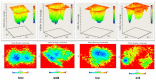
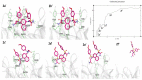
In this study, we investigate the atomistic details of Keap1-Nrf2 inhibitors by in-depth modeling techniques, including molecular dynamics (MD) simulations, and the path-based free energy method of umbrella sampling (US). The protein-protein interaction (PPI) of Keap1-Nrf2 is implicated in several neurodegenerative diseases like cancer, diabetes, and cardiomyopathy. A better understanding of the five sub-pocket binding sites for Nrf2 (ETGE and DLG motifs) inside the Kelch domain would expedite the inhibitor design process. We selected four protein-ligand complexes with distinct co-crystal ligands and binding occupancies inside the Nrf2 binding site. We performed 100 ns of MD simulation for each complex and analyzed the trajectories. From the results, it is evident that one ligand (1VV) has flipped inside the binding pocket, whereas the remaining three were stable. We found that Coulombic (Arg483, Arg415, Ser363, Ser508, and Ser602) and Lennard-Jones (Tyr525, Tyr334, and Tyr572) interactions played a significant role in complex stability. The obtained binding free energy values from US simulations were consistent with the potencies of simulated ligands. US simulation highlight the importance of basic and aromatic residues in the binding pocket. A detailed description of the dissociation process brings valuable insight into the interaction of the four selected protein-ligand complexes, which could help in the future to design more potent PPI inhibitors.
Keywords: Keap1-NRF2 inhibitors; MD simulations; PPI inhibition; US simulation; binding free energy; molecular modeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Marcotte D., Zeng W., Hus J.C., McKenzie A., Hession C., Jin P., Bergeron C., Lugovskoy A., Enyedy I., Cuervo H., et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorgan. Med. Chem. 2013;21:4011–4019. doi: 10.1016/j.bmc.2013.04.019. - DOI - PubMed
-
- Bertrand H.C., Schaap M., Baird L., Georgakopoulos N.D., Fowkes A., Thiollier C., Kachi H., Dinkova-Kostova A.T., Wells G. Design, synthesis, and evaluation of triazole derivatives that induce Nrf2 dependent gene products and inhibit the Keap1–Nrf2 protein–protein interaction. J. Med. Chem. 2015;58:7186–7194. doi: 10.1021/acs.jmedchem.5b00602. - DOI - PubMed
-
- Yasuda D., Nakajima M., Yuasa A., Obata R., Takahashi K., Ohe T., Ichimura Y., Komatsu M., Yamamoto M., Imamura R., et al. Synthesis of Keap1-phosphorylated p62 and Keap1-Nrf2 protein-protein interaction inhibitors and their inhibitory activity. Bioorganic Med. Chem. Lett. 2016;26:5956–5959. doi: 10.1016/j.bmcl.2016.10.083. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

