Application of CRISPR genetic screens to investigate neurological diseases
- PMID: 31727120
- PMCID: PMC6857349
- DOI: 10.1186/s13024-019-0343-3
Application of CRISPR genetic screens to investigate neurological diseases
Abstract
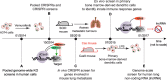
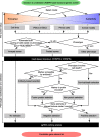
The adoption of CRISPR-Cas9 technology for functional genetic screens has been a transformative advance. Due to its modular nature, this technology can be customized to address a myriad of questions. To date, pooled, genome-scale studies have uncovered genes responsible for survival, proliferation, drug resistance, viral susceptibility, and many other functions. The technology has even been applied to the functional interrogation of the non-coding genome. However, applications of this technology to neurological diseases remain scarce. This shortfall motivated the assembly of a review that will hopefully help researchers moving in this direction find their footing. The emphasis here will be on design considerations and concepts underlying this methodology. We will highlight groundbreaking studies in the CRISPR-Cas9 functional genetics field and discuss strengths and limitations of this technology for neurological disease applications. Finally, we will provide practical guidance on navigating the many choices that need to be made when implementing a CRISPR-Cas9 functional genetic screen for the study of neurological diseases.
Keywords: CRISIPR KO; CRISPR-Cas9; CRISPRa; CRISPRi; Functional genetics; Marker selection screens; Neurodegenerative diseases; Neurological diseases; Survival screens; sgRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

