Disparate responses to salinity across species and organizational levels in anchialine shrimps
- PMID: 31727759
- PMCID: PMC6955204
- DOI: 10.1242/jeb.211920
Disparate responses to salinity across species and organizational levels in anchialine shrimps
Abstract
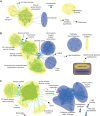
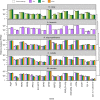
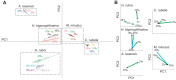
Environmentally induced plasticity in gene expression is one of the underlying mechanisms of adaptation to habitats with variable environments. For example, euryhaline crustaceans show predictable changes in the expression of ion-transporter genes during salinity transfers, although studies have typically been limited to specific genes, taxa and ecosystems of interest. Here, we investigated responses to salinity change at multiple organizational levels in five species of shrimp representing at least three independent invasions of the anchialine ecosystem, defined as habitats with marine and freshwater influences with spatial and temporal fluctuations in salinity. Although all five species were generally strong osmoregulators, salinity-induced changes in gill physiology and gene expression were highly species specific. While some species exhibited patterns similar to those of previously studied euryhaline crustaceans, instances of distinct and atypical patterns were recovered from closely related species. Species-specific patterns were found when examining: (1) numbers and identities of differentially expressed genes, (2) salinity-induced expression of genes predicted a priori to play a role in osmoregulation, and (3) salinity-induced expression of orthologs shared among all species. Notably, ion transport genes were unchanged in the atyid Halocaridina rubra while genes normally associated with vision and light perception were among those most highly upregulated. Potential reasons for species-specific patterns are discussed, including variation among anchialine habitats in salinity regimes and divergent evolution in anchialine taxa. Underexplored mechanisms of osmoregulation in crustaceans revealed here by the application of transcriptomic approaches to ecologically and taxonomically understudied systems are also explored.
Keywords: Acclimation; Atyidae; Euryhalinity; RNA-Seq.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Anker A. (2010). Metabetaeus Borradaile, 1899 revisited, with description of a new marine species from French Polynesia (Crustacea: Decapoda: Alpheidae). Zootaxa 2552, 37-54. 10.11646/zootaxa.2552.1.2 - DOI
-
- Cameron J. N. (1978). NaCl balance in blue crabs, Callinectes sapidus, in fresh water. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 123, 127-135. 10.1007/BF00687840 - DOI
-
- Chen K., Li E. C., Gan L., Wang X. D., Xu C., Lin H. Z., Qin J. G. and Chen L. Q. (2014). Growth and lipid metabolism of the pacific white shrimp Litopenaeus vannamei at different salinities. J. Shellfish Res. 33, 825-832. 10.2983/035.033.0317 - DOI
-
- Chen K., Li E. C., Li T. Y., Xu C., Wang X. D., Lin H. Z., Qin J. G. and Chen L. Q. (2015). Transcriptome and molecular pathway analysis of the hepatopancreas in the pacific white shrimp Litopenaeus vannamei under chronic low-salinity stress. PLoS ONE 10, e0131503 10.1371/journal.pone.0131503 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

