Cellular RNA surveillance in health and disease
- PMID: 31727827
- PMCID: PMC6938259
- DOI: 10.1126/science.aax2957
Cellular RNA surveillance in health and disease
Abstract
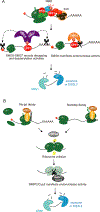
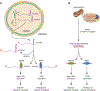
The numerous quality control pathways that target defective ribonucleic acids (RNAs) for degradation play key roles in shaping mammalian transcriptomes and preventing disease. These pathways monitor most steps in the biogenesis of both noncoding RNAs (ncRNAs) and protein-coding messenger RNAs (mRNAs), degrading ncRNAs that fail to form functional complexes with one or more proteins and eliminating mRNAs that encode abnormal, potentially toxic proteins. Mutations in components of diverse RNA surveillance pathways manifest as disease. Some mutations are characterized by increased interferon production, suggesting that a major role of these pathways is to prevent aberrant cellular RNAs from being recognized as "non-self." Other mutations are common in cancer, or result in developmental defects, revealing the importance of RNA surveillance to cell and organismal function.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


References
-
- Lubas M et al., Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

