Fecal microbiome signatures of pancreatic cancer patients
- PMID: 31727922
- PMCID: PMC6856127
- DOI: 10.1038/s41598-019-53041-4
Fecal microbiome signatures of pancreatic cancer patients
Abstract
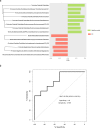
Pancreatic cancer (PC) is a leading cause of cancer-related death in developed countries, and since most patients have incurable disease at the time of diagnosis, developing a screening method for early detection is of high priority. Due to its metabolic importance, alterations in pancreatic functions may affect the composition of the gut microbiota, potentially yielding biomarkers for PC. However, the usefulness of these biomarkers may be limited if they are specific for advanced stages of disease, which may involve comorbidities such as biliary obstruction or diabetes. In this study we analyzed the fecal microbiota of 30 patients with pancreatic adenocarcinoma, 6 patients with pre-cancerous lesions, 13 healthy subjects and 16 with non-alcoholic fatty liver disease, using amplicon sequencing of the bacterial 16S rRNA gene. Fourteen bacterial features discriminated between PC and controls, and several were shared with findings from a recent Chinese cohort. A Random Forest model based on the microbiota classified PC and control samples with an AUC of 82.5%. However, inter-subject variability was high, and only a small part of the PC-associated microbial signals were also observed in patients with pre-cancerous pancreatic lesions, implying that microbiome-based early detection of such lesions will be challenging.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

